Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
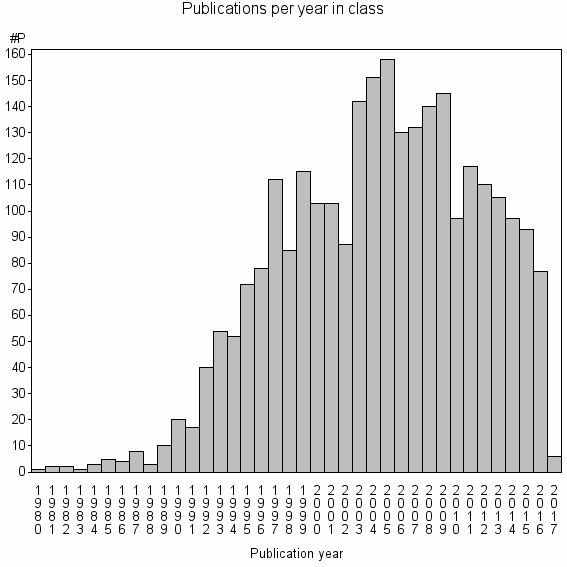
| 1409 | 2677 | 43.2 | 80% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 1409 | 1 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//THREADING | 2677 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN STRUCTURE PREDICTION | authKW | 1974727 | 14% | 45% | 383 |
| 2 | FOLD RECOGNITION | authKW | 869742 | 5% | 58% | 131 |
| 3 | THREADING | authKW | 763746 | 4% | 56% | 120 |
| 4 | CASP | authKW | 658891 | 3% | 66% | 87 |
| 5 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | journal | 545645 | 21% | 9% | 552 |
| 6 | KNOWLEDGE BASED POTENTIALS | authKW | 370915 | 2% | 59% | 55 |
| 7 | STATISTICAL POTENTIALS | authKW | 350019 | 2% | 57% | 54 |
| 8 | PROTEIN THREADING | authKW | 289793 | 1% | 71% | 36 |
| 9 | LOOP MODELING | authKW | 289083 | 1% | 65% | 39 |
| 10 | STRUCTURE PREDICTION | authKW | 241114 | 5% | 17% | 123 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biophysics | 20645 | 32% | 0% | 857 |
| 2 | Mathematical & Computational Biology | 16709 | 15% | 0% | 393 |
| 3 | Biochemistry & Molecular Biology | 13419 | 58% | 0% | 1555 |
| 4 | Biochemical Research Methods | 7128 | 18% | 0% | 476 |
| 5 | Biotechnology & Applied Microbiology | 2050 | 13% | 0% | 346 |
| 6 | Computer Science, Interdisciplinary Applications | 1174 | 6% | 0% | 167 |
| 7 | Physics, Atomic, Molecular & Chemical | 196 | 5% | 0% | 126 |
| 8 | Chemistry, Multidisciplinary | 88 | 7% | 0% | 187 |
| 9 | Chemistry, Physical | 71 | 7% | 0% | 185 |
| 10 | Statistics & Probability | 61 | 2% | 0% | 46 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BAKER CHEM CHEM BIOL | 233055 | 3% | 27% | 77 |
| 2 | THEORY BIOPOLYMERS | 82761 | 1% | 48% | 15 |
| 3 | PELS FAMILY BIOCHEM STRUCT BIOL | 74118 | 0% | 50% | 13 |
| 4 | COMPUTAT PROTEOM BIOPHYS | 61578 | 0% | 60% | 9 |
| 5 | PL MOL ENGN | 57298 | 1% | 36% | 14 |
| 6 | STUDY SYST BIOL | 52405 | 1% | 23% | 20 |
| 7 | SEAVER FDN BIOINFORMAT | 52129 | 0% | 57% | 8 |
| 8 | EBGM | 51829 | 0% | 45% | 10 |
| 9 | C BOND LIFE SCI | 49574 | 0% | 43% | 10 |
| 10 | BIOINFORMAT MOL DESIGN TECHNOL INNOVAT | 42981 | 0% | 54% | 7 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 545645 | 21% | 9% | 552 |
| 2 | FOLDING & DESIGN | 31147 | 1% | 12% | 22 |
| 3 | PROTEIN SCIENCE | 31069 | 5% | 2% | 131 |
| 4 | BIOINFORMATICS | 30051 | 6% | 2% | 168 |
| 5 | PROTEIN ENGINEERING | 29040 | 3% | 3% | 76 |
| 6 | BMC STRUCTURAL BIOLOGY | 26252 | 1% | 7% | 34 |
| 7 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 14310 | 3% | 1% | 88 |
| 8 | BMC BIOINFORMATICS | 12731 | 3% | 1% | 92 |
| 9 | PROTEINS-STRUCTURE FUNCTION AND GENETICS | 9236 | 1% | 4% | 19 |
| 10 | JOURNAL OF MOLECULAR BIOLOGY | 8753 | 5% | 1% | 135 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN STRUCTURE PREDICTION | 1974727 | 14% | 45% | 383 | Search PROTEIN+STRUCTURE+PREDICTION | Search PROTEIN+STRUCTURE+PREDICTION |
| 2 | FOLD RECOGNITION | 869742 | 5% | 58% | 131 | Search FOLD+RECOGNITION | Search FOLD+RECOGNITION |
| 3 | THREADING | 763746 | 4% | 56% | 120 | Search THREADING | Search THREADING |
| 4 | CASP | 658891 | 3% | 66% | 87 | Search CASP | Search CASP |
| 5 | KNOWLEDGE BASED POTENTIALS | 370915 | 2% | 59% | 55 | Search KNOWLEDGE+BASED+POTENTIALS | Search KNOWLEDGE+BASED+POTENTIALS |
| 6 | STATISTICAL POTENTIALS | 350019 | 2% | 57% | 54 | Search STATISTICAL+POTENTIALS | Search STATISTICAL+POTENTIALS |
| 7 | PROTEIN THREADING | 289793 | 1% | 71% | 36 | Search PROTEIN+THREADING | Search PROTEIN+THREADING |
| 8 | LOOP MODELING | 289083 | 1% | 65% | 39 | Search LOOP+MODELING | Search LOOP+MODELING |
| 9 | STRUCTURE PREDICTION | 241114 | 5% | 17% | 123 | Search STRUCTURE+PREDICTION | Search STRUCTURE+PREDICTION |
| 10 | LOOP PREDICTION | 211896 | 1% | 77% | 24 | Search LOOP+PREDICTION | Search LOOP+PREDICTION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ROY, A , KUCUKURAL, A , ZHANG, Y , (2010) I-TASSER: A UNIFIED PLATFORM FOR AUTOMATED PROTEIN STRUCTURE AND FUNCTION PREDICTION.NATURE PROTOCOLS. VOL. 5. ISSUE 4. P. 725-738 | 46 | 74% | 1865 |
| 2 | SALI, A , SHEN, MY , (2006) STATISTICAL POTENTIAL FOR ASSESSMENT AND PREDICTION OF PROTEIN STRUCTURES.PROTEIN SCIENCE. VOL. 15. ISSUE 11. P. 2507 -2524 | 73 | 81% | 728 |
| 3 | DORN, M , SILVA, MBE , BURIOL, LS , LAMB, LC , (2014) THREE-DIMENSIONAL PROTEIN STRUCTURE PREDICTION: METHODS AND COMPUTATIONAL STRATEGIES.COMPUTATIONAL BIOLOGY AND CHEMISTRY. VOL. 53. ISSUE . P. 251 -276 | 173 | 50% | 7 |
| 4 | BIASINI, M , BIENERT, S , WATERHOUSE, A , ARNOLD, K , STUDER, G , SCHMIDT, T , KIEFER, F , CASSARINO, TG , BERTONI, M , BORDOLI, L , ET AL (2014) SWISS-MODEL: MODELLING PROTEIN TERTIARY AND QUATERNARY STRUCTURE USING EVOLUTIONARY INFORMATION.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE W1. P. W252 -W258 | 24 | 89% | 663 |
| 5 | BENKERT, P , TOSATTO, SCE , SCHOMBURG, D , (2008) QMEAN: A COMPREHENSIVE SCORING FUNCTION FOR MODEL QUALITY ASSESSMENT.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 71. ISSUE 1. P. 261 -277 | 55 | 83% | 279 |
| 6 | JOSEPH, AP , DE BREVERN, AG , (2014) FROM LOCAL STRUCTURE TO A GLOBAL FRAMEWORK: RECOGNITION OF PROTEIN FOLDS.JOURNAL OF THE ROYAL SOCIETY INTERFACE. VOL. 11. ISSUE 95. P. - | 118 | 52% | 1 |
| 7 | QU, XT , SWANSON, R , DAY, R , TSAI, J , (2009) A GUIDE TO TEMPLATE BASED STRUCTURE PREDICTION.CURRENT PROTEIN & PEPTIDE SCIENCE. VOL. 10. ISSUE 3. P. 270 -285 | 114 | 57% | 20 |
| 8 | JAYARAM, B , DHINGRA, P , (2012) TOWARDS CREATING COMPLETE PROTEOMIC STRUCTURAL DATABASES OF WHOLE ORGANISMS.CURRENT BIOINFORMATICS. VOL. 7. ISSUE 4. P. 424-435 | 94 | 66% | 1 |
| 9 | MELO, F , SANCHEZ, R , SALI, A , (2002) STATISTICAL POTENTIALS FOR FOLD ASSESSMENT.PROTEIN SCIENCE. VOL. 11. ISSUE 2. P. 430 -448 | 78 | 79% | 161 |
| 10 | FLOUDAS, CA , (2007) COMPUTATIONAL METHODS IN PROTEIN STRUCTURE PREDICTION.BIOTECHNOLOGY AND BIOENGINEERING. VOL. 97. ISSUE 2. P. 207-213 | 67 | 86% | 53 |
Classes with closest relation at Level 1 |