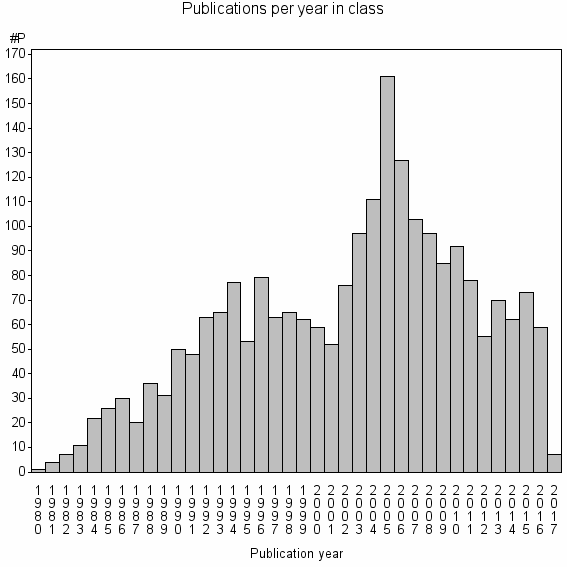
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 2338 | 2277 | 28.7 | 66% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 2338 | 1 | MULTIPLE SEQUENCE ALIGNMENT//SEQUENCE ALIGNMENT//COMPUTER APPLICATIONS IN THE BIOSCIENCES | 2277 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MULTIPLE SEQUENCE ALIGNMENT | authKW | 589545 | 5% | 36% | 123 |
| 2 | SEQUENCE ALIGNMENT | authKW | 450590 | 6% | 23% | 146 |
| 3 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | journal | 377359 | 7% | 18% | 160 |
| 4 | SPACED SEEDS | authKW | 193748 | 1% | 85% | 17 |
| 5 | LOCAL ALIGNMENT | authKW | 151600 | 1% | 54% | 21 |
| 6 | STATISTICAL ALIGNMENT | authKW | 134086 | 0% | 100% | 10 |
| 7 | SMITH WATERMAN | authKW | 131160 | 1% | 65% | 15 |
| 8 | T COFFEE | authKW | 121894 | 0% | 91% | 10 |
| 9 | JOURNAL OF COMPUTATIONAL BIOLOGY | journal | 89407 | 4% | 7% | 102 |
| 10 | HOMOLOGY SEARCH | authKW | 87963 | 1% | 25% | 26 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 65124 | 31% | 1% | 707 |
| 2 | Biochemical Research Methods | 18479 | 30% | 0% | 689 |
| 3 | Biotechnology & Applied Microbiology | 9832 | 28% | 0% | 647 |
| 4 | Computer Science, Interdisciplinary Applications | 8458 | 17% | 0% | 383 |
| 5 | Computer Science, Theory & Methods | 3361 | 11% | 0% | 253 |
| 6 | Statistics & Probability | 2468 | 8% | 0% | 193 |
| 7 | Biochemistry & Molecular Biology | 1893 | 27% | 0% | 613 |
| 8 | Evolutionary Biology | 1832 | 6% | 0% | 143 |
| 9 | Genetics & Heredity | 565 | 8% | 0% | 180 |
| 10 | Computer Science, Artificial Intelligence | 564 | 5% | 0% | 107 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | INTERDISCIPLINARY STUDIES STRUCT FORMAT | 47885 | 0% | 71% | 5 |
| 2 | DEP LENGUAJES CIENCIAS COMPUTAC | 30168 | 0% | 75% | 3 |
| 3 | BIOL FR 36 | 26817 | 0% | 100% | 2 |
| 4 | PHYSIOL CELLULAIRE VEGETALE CEA GRENOBLE | 26817 | 0% | 100% | 2 |
| 5 | SYMBIOSE TEAM PROJECT | 26817 | 0% | 100% | 2 |
| 6 | COMPARAT BIOINFORMAT GRP | 26813 | 0% | 50% | 4 |
| 7 | UFR SES | 20110 | 0% | 50% | 3 |
| 8 | EDUC TECHNOL COMP | 17877 | 0% | 67% | 2 |
| 9 | INFORMAT INTERDISZIPLINA | 17877 | 0% | 67% | 2 |
| 10 | BIOTECHNOL INFORMAT | 16024 | 2% | 2% | 51 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 377359 | 7% | 18% | 160 |
| 2 | JOURNAL OF COMPUTATIONAL BIOLOGY | 89407 | 4% | 7% | 102 |
| 3 | BIOINFORMATICS | 85045 | 11% | 2% | 260 |
| 4 | BMC BIOINFORMATICS | 39527 | 7% | 2% | 149 |
| 5 | ALGORITHMS FOR MOLECULAR BIOLOGY | 12099 | 1% | 6% | 16 |
| 6 | NUCLEIC ACIDS RESEARCH | 10450 | 8% | 0% | 177 |
| 7 | GENE-COMBIS | 10052 | 0% | 25% | 3 |
| 8 | IEEE-ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS | 7971 | 1% | 2% | 26 |
| 9 | BULLETIN OF MATHEMATICAL BIOLOGY | 6982 | 2% | 1% | 36 |
| 10 | EVOLUTIONARY BIOINFORMATICS | 6107 | 1% | 4% | 12 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MULTIPLE SEQUENCE ALIGNMENT | 589545 | 5% | 36% | 123 | Search MULTIPLE+SEQUENCE+ALIGNMENT | Search MULTIPLE+SEQUENCE+ALIGNMENT |
| 2 | SEQUENCE ALIGNMENT | 450590 | 6% | 23% | 146 | Search SEQUENCE+ALIGNMENT | Search SEQUENCE+ALIGNMENT |
| 3 | SPACED SEEDS | 193748 | 1% | 85% | 17 | Search SPACED+SEEDS | Search SPACED+SEEDS |
| 4 | LOCAL ALIGNMENT | 151600 | 1% | 54% | 21 | Search LOCAL+ALIGNMENT | Search LOCAL+ALIGNMENT |
| 5 | STATISTICAL ALIGNMENT | 134086 | 0% | 100% | 10 | Search STATISTICAL+ALIGNMENT | Search STATISTICAL+ALIGNMENT |
| 6 | SMITH WATERMAN | 131160 | 1% | 65% | 15 | Search SMITH+WATERMAN | Search SMITH+WATERMAN |
| 7 | T COFFEE | 121894 | 0% | 91% | 10 | Search T+COFFEE | Search T+COFFEE |
| 8 | HOMOLOGY SEARCH | 87963 | 1% | 25% | 26 | Search HOMOLOGY+SEARCH | Search HOMOLOGY+SEARCH |
| 9 | SMITH WATERMAN ALGORITHM | 87754 | 1% | 55% | 12 | Search SMITH+WATERMAN+ALGORITHM | Search SMITH+WATERMAN+ALGORITHM |
| 10 | MULTIPLE ALIGNMENT | 85703 | 1% | 24% | 27 | Search MULTIPLE+ALIGNMENT | Search MULTIPLE+ALIGNMENT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ALTSCHUL, SF , MADDEN, TL , SCHAFFER, AA , ZHANG, JH , ZHANG, Z , MILLER, W , LIPMAN, DJ , (1997) GAPPED BLAST AND PSI-BLAST: A NEW GENERATION OF PROTEIN DATABASE SEARCH PROGRAMS.NUCLEIC ACIDS RESEARCH. VOL. 25. ISSUE 17. P. 3389-3402 | 37 | 61% | 41839 |
| 2 | MORRISON, DA , (2006) MULTIPLE SEQUENCE ALIGNMENT FOR PHYLOGENETIC PURPOSES.AUSTRALIAN SYSTEMATIC BOTANY. VOL. 19. ISSUE 6. P. 479 -539 | 158 | 49% | 73 |
| 3 | ALTSCHUL, SF , GISH, W , MILLER, W , MYERS, EW , LIPMAN, DJ , (1990) BASIC LOCAL ALIGNMENT SEARCH TOOL.JOURNAL OF MOLECULAR BIOLOGY. VOL. 215. ISSUE 3. P. 403-410 | 10 | 83% | 45094 |
| 4 | HERMAN, JL , NOVAK, A , LYNGSO, R , SZABO, A , MIKLOS, I , HEIN, J , (2015) EFFICIENT REPRESENTATION OF UNCERTAINTY IN MULTIPLE SEQUENCE ALIGNMENTS USING DIRECTED ACYCLIC GRAPHS.BMC BIOINFORMATICS. VOL. 16. ISSUE . P. - | 67 | 66% | 4 |
| 5 | SIEVERS, F , WILM, A , DINEEN, D , GIBSON, TJ , KARPLUS, K , LI, WZ , LOPEZ, R , MCWILLIAM, H , REMMERT, M , SODING, J , ET AL (2011) FAST, SCALABLE GENERATION OF HIGH-QUALITY PROTEIN MULTIPLE SEQUENCE ALIGNMENTS USING CLUSTAL OMEGA.MOLECULAR SYSTEMS BIOLOGY. VOL. 7. ISSUE . P. - | 17 | 74% | 1990 |
| 6 | EDGAR, RC , (2004) MUSCLE: MULTIPLE SEQUENCE ALIGNMENT WITH HIGH ACCURACY AND HIGH THROUGHPUT.NUCLEIC ACIDS RESEARCH. VOL. 32. ISSUE 5. P. 1792-1797 | 17 | 53% | 12512 |
| 7 | GOTOH, O , (1999) MULTIPLE SEQUENCE ALIGNMENT: ALGORITHMS AND APPLICATIONS.ADVANCES IN BIOPHYSICS, VOL 36, 1999. VOL. 36. ISSUE . P. 159 -206 | 97 | 66% | 50 |
| 8 | KATOH, K , MISAWA, K , KUMA, K , MIYATA, T , (2002) MAFFT: A NOVEL METHOD FOR RAPID MULTIPLE SEQUENCE ALIGNMENT BASED ON FAST FOURIER TRANSFORM.NUCLEIC ACIDS RESEARCH. VOL. 30. ISSUE 14. P. 3059 -3066 | 22 | 79% | 3341 |
| 9 | EZAWA, K , (2016) CHARACTERIZATION OF MULTIPLE SEQUENCE ALIGNMENT ERRORS USING COMPLETE-LIKELIHOOD SCORE AND POSITION-SHIFT MAP.BMC BIOINFORMATICS. VOL. 17. ISSUE . P. - | 51 | 78% | 0 |
| 10 | KATOH, K , STANDLEY, DM , (2016) A SIMPLE METHOD TO CONTROL OVER-ALIGNMENT IN THE MAFFT MULTIPLE SEQUENCE ALIGNMENT PROGRAM.BIOINFORMATICS. VOL. 32. ISSUE 13. P. 1933 -1942 | 38 | 76% | 5 |
Classes with closest relation at Level 1 |