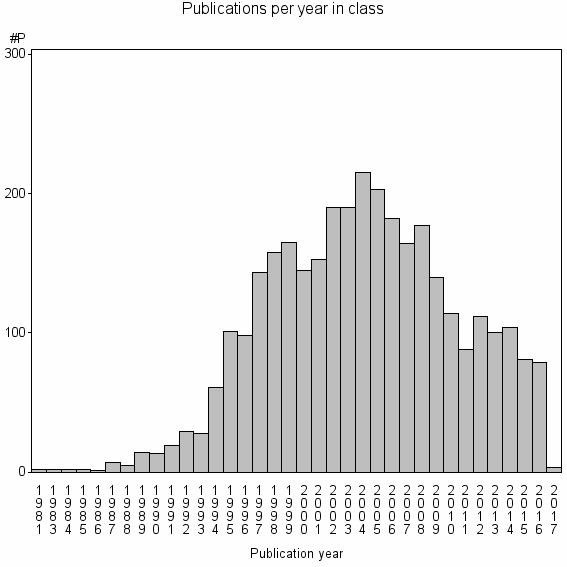
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 573 | 3290 | 48.3 | 85% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 260 | 2 | PROTEIN FOLDING//JOURNAL OF CHEMICAL THEORY AND COMPUTATION//JOURNAL OF COMPUTATIONAL CHEMISTRY | 19582 |
| 573 | 1 | PROTEIN FOLDING//PHI VALUE//FOLDING RATE | 3290 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | authKW | 782823 | 21% | 12% | 703 |
| 2 | PHI VALUE | authKW | 460348 | 2% | 79% | 63 |
| 3 | FOLDING RATE | authKW | 288024 | 1% | 80% | 39 |
| 4 | CONTACT ORDER | authKW | 270272 | 1% | 79% | 37 |
| 5 | PHI VALUE ANALYSIS | authKW | 239666 | 1% | 70% | 37 |
| 6 | FOLDING KINETICS | authKW | 233935 | 2% | 38% | 67 |
| 7 | FOLDING NUCLEUS | authKW | 233178 | 1% | 74% | 34 |
| 8 | PROTEIN FOLDING KINETICS | authKW | 174200 | 1% | 57% | 33 |
| 9 | FOLDING PATHWAY | authKW | 154863 | 1% | 35% | 48 |
| 10 | TRANSITION STATE ENSEMBLE | authKW | 154859 | 1% | 76% | 22 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 16149 | 58% | 0% | 1893 |
| 2 | Biophysics | 9100 | 20% | 0% | 647 |
| 3 | Physics, Atomic, Molecular & Chemical | 2397 | 12% | 0% | 386 |
| 4 | Chemistry, Physical | 1447 | 17% | 0% | 547 |
| 5 | Physics, Mathematical | 1028 | 6% | 0% | 184 |
| 6 | Physics, Fluids & Plasmas | 576 | 4% | 0% | 124 |
| 7 | Mathematical & Computational Biology | 520 | 3% | 0% | 86 |
| 8 | Physics, Multidisciplinary | 370 | 7% | 0% | 217 |
| 9 | Cell Biology | 142 | 6% | 0% | 182 |
| 10 | Biochemical Research Methods | 120 | 3% | 0% | 99 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMAT TELEMED | 91773 | 1% | 34% | 29 |
| 2 | THEORET BIOL PHYS | 87758 | 3% | 10% | 91 |
| 3 | PROGRAM BIOCHEM STRUCT BIOL | 84779 | 1% | 26% | 35 |
| 4 | MRC PROT ENGN | 38653 | 0% | 42% | 10 |
| 5 | BIOMOL STRUCT DESIGN PROGRAM | 32969 | 0% | 22% | 16 |
| 6 | PROGRAM BIOPHYS | 31766 | 1% | 8% | 45 |
| 7 | CHIM BIOPHYS | 31126 | 1% | 12% | 27 |
| 8 | CAMBRIDGE PROT ENGN | 30139 | 0% | 25% | 13 |
| 9 | PROT ENGN | 28747 | 2% | 5% | 66 |
| 10 | GLOBAL STRATEG ANALYT UNIT | 27838 | 0% | 100% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | FOLDING & DESIGN | 71713 | 1% | 21% | 37 |
| 2 | JOURNAL OF MOLECULAR BIOLOGY | 71560 | 13% | 2% | 424 |
| 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 43352 | 5% | 3% | 173 |
| 4 | PROTEIN SCIENCE | 27204 | 4% | 2% | 136 |
| 5 | NATURE STRUCTURAL BIOLOGY | 24784 | 2% | 4% | 65 |
| 6 | CURRENT OPINION IN STRUCTURAL BIOLOGY | 11804 | 2% | 2% | 54 |
| 7 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 9906 | 10% | 0% | 323 |
| 8 | BIOCHEMISTRY | 9182 | 7% | 0% | 233 |
| 9 | JOURNAL OF CHEMICAL PHYSICS | 6364 | 8% | 0% | 249 |
| 10 | BIOPHYSICAL JOURNAL | 6242 | 4% | 1% | 116 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN FOLDING | 782823 | 21% | 12% | 703 | Search PROTEIN+FOLDING | Search PROTEIN+FOLDING |
| 2 | PHI VALUE | 460348 | 2% | 79% | 63 | Search PHI+VALUE | Search PHI+VALUE |
| 3 | FOLDING RATE | 288024 | 1% | 80% | 39 | Search FOLDING+RATE | Search FOLDING+RATE |
| 4 | CONTACT ORDER | 270272 | 1% | 79% | 37 | Search CONTACT+ORDER | Search CONTACT+ORDER |
| 5 | PHI VALUE ANALYSIS | 239666 | 1% | 70% | 37 | Search PHI+VALUE+ANALYSIS | Search PHI+VALUE+ANALYSIS |
| 6 | FOLDING KINETICS | 233935 | 2% | 38% | 67 | Search FOLDING+KINETICS | Search FOLDING+KINETICS |
| 7 | FOLDING NUCLEUS | 233178 | 1% | 74% | 34 | Search FOLDING+NUCLEUS | Search FOLDING+NUCLEUS |
| 8 | PROTEIN FOLDING KINETICS | 174200 | 1% | 57% | 33 | Search PROTEIN+FOLDING+KINETICS | Search PROTEIN+FOLDING+KINETICS |
| 9 | FOLDING PATHWAY | 154863 | 1% | 35% | 48 | Search FOLDING+PATHWAY | Search FOLDING+PATHWAY |
| 10 | TRANSITION STATE ENSEMBLE | 154859 | 1% | 76% | 22 | Search TRANSITION+STATE+ENSEMBLE | Search TRANSITION+STATE+ENSEMBLE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SHAKHNOVICH, E , (2006) PROTEIN FOLDING THERMODYNAMICS AND DYNAMICS: WHERE PHYSICS, CHEMISTRY, AND BIOLOGY MEET.CHEMICAL REVIEWS. VOL. 106. ISSUE 5. P. 1559-1588 | 176 | 74% | 208 |
| 2 | SOSNICK, TR , BARRICK, D , (2011) THE FOLDING OF SINGLE DOMAIN PROTEINS - HAVE WE REACHED A CONSENSUS?.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 21. ISSUE 1. P. 12-24 | 144 | 75% | 70 |
| 3 | OLIVEBERG, M , WOLYNES, PG , (2005) THE EXPERIMENTAL SURVEY OF PROTEIN-FOLDING ENERGY LANDSCAPES.QUARTERLY REVIEWS OF BIOPHYSICS. VOL. 38. ISSUE 3. P. 245 -288 | 132 | 84% | 138 |
| 4 | WEIKL, TR , (2008) LOOP-CLOSURE PRINCIPLES IN PROTEIN FOLDING.ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS. VOL. 469. ISSUE 1. P. 67-75 | 126 | 89% | 13 |
| 5 | MALHOTRA, P , UDGAONKAR, JB , (2016) HOW COOPERATIVE ARE PROTEIN FOLDING AND UNFOLDING TRANSITIONS?.PROTEIN SCIENCE. VOL. 25. ISSUE 11. P. 1924 -1941 | 131 | 63% | 0 |
| 6 | PLOTKIN, SS , ONUCHIC, JN , (2002) UNDERSTANDING PROTEIN FOLDING WITH ENERGY LANDSCAPE THEORY - PART I: BASIC CONCEPTS.QUARTERLY REVIEWS OF BIOPHYSICS. VOL. 35. ISSUE 2. P. 111 -167 | 143 | 65% | 122 |
| 7 | DILL, KA , OZKAN, SB , SHELL, MS , WEIKL, TR , (2008) THE PROTEIN FOLDING PROBLEM.ANNUAL REVIEW OF BIOPHYSICS. VOL. 37. ISSUE . P. 289 -316 | 107 | 48% | 392 |
| 8 | SINHA, KK , UDGAONKAR, JB , (2009) EARLY EVENTS IN PROTEIN FOLDING.CURRENT SCIENCE. VOL. 96. ISSUE 8. P. 1053 -1070 | 130 | 64% | 9 |
| 9 | JHA, SK , UDGAONKAR, JB , (2010) FREE ENERGY BARRIERS IN PROTEIN FOLDING AND UNFOLDING REACTIONS.CURRENT SCIENCE. VOL. 99. ISSUE 4. P. 457-475 | 129 | 64% | 6 |
| 10 | MIRNY, L , SHAKHNOVICH, E , (2001) PROTEIN FOLDING THEORY: FROM LATTICE TO ALL-ATOM MODELS.ANNUAL REVIEW OF BIOPHYSICS AND BIOMOLECULAR STRUCTURE. VOL. 30. ISSUE . P. 361 -396 | 97 | 84% | 231 |
Classes with closest relation at Level 1 |