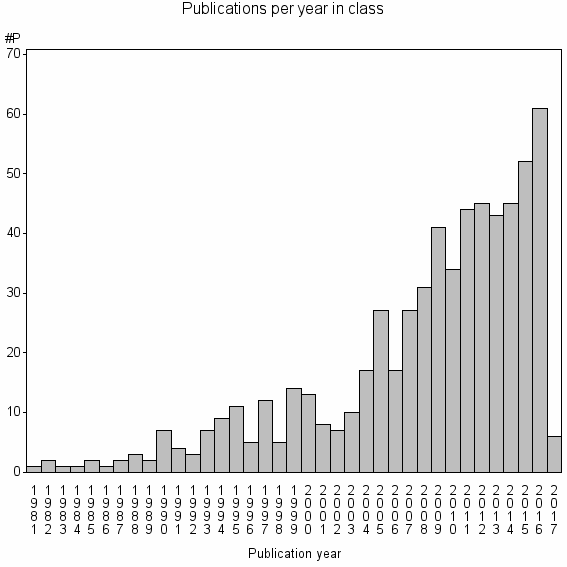
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 17055 | 620 | 37.0 | 74% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 17055 | 1 | CORRELATED MUTATIONS//DIRECT COUPLING ANALYSIS//INVERSE ISING PROBLEM | 620 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CORRELATED MUTATIONS | authKW | 395345 | 3% | 47% | 17 |
| 2 | DIRECT COUPLING ANALYSIS | authKW | 253281 | 1% | 86% | 6 |
| 3 | INVERSE ISING PROBLEM | authKW | 246247 | 1% | 100% | 5 |
| 4 | CONTACT PREDICTION | authKW | 246237 | 2% | 50% | 10 |
| 5 | CONTACT MAP | authKW | 222209 | 3% | 24% | 19 |
| 6 | CONTACT MAP PREDICTION | authKW | 175888 | 1% | 71% | 5 |
| 7 | PROTEIN CONTACT MAP | authKW | 157596 | 1% | 80% | 4 |
| 8 | RESIDUE RESIDUE CONTACT | authKW | 157596 | 1% | 80% | 4 |
| 9 | COEVOLVING RESIDUES | authKW | 147748 | 0% | 100% | 3 |
| 10 | CORRELATED SUBSTITUTIONS | authKW | 147748 | 0% | 100% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 9850 | 23% | 0% | 144 |
| 2 | Biochemical Research Methods | 3159 | 24% | 0% | 150 |
| 3 | Physics, Mathematical | 1125 | 12% | 0% | 77 |
| 4 | Biotechnology & Applied Microbiology | 891 | 17% | 0% | 106 |
| 5 | Biochemistry & Molecular Biology | 884 | 34% | 0% | 208 |
| 6 | Biophysics | 544 | 12% | 0% | 72 |
| 7 | Computer Science, Artificial Intelligence | 352 | 7% | 0% | 42 |
| 8 | Physics, Fluids & Plasmas | 209 | 5% | 0% | 31 |
| 9 | Evolutionary Biology | 189 | 4% | 0% | 25 |
| 10 | Computer Science, Interdisciplinary Applications | 98 | 4% | 0% | 25 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOM MICROORGANISMES | 105054 | 1% | 27% | 8 |
| 2 | BIOL COMPUTAT QUANTITAT BIOL PARIS SEINE | 98499 | 0% | 100% | 2 |
| 3 | UMR 7238 | 86386 | 2% | 18% | 10 |
| 4 | EVOLUTIONARY GENET BIOINFORMAT | 65660 | 1% | 33% | 4 |
| 5 | TAO INRIA | 55402 | 0% | 38% | 3 |
| 6 | ABIOT ST SGRP INTEGRAT SYST BIOL | 49249 | 0% | 100% | 1 |
| 7 | ACCESS LINNAEUS KTH | 49249 | 0% | 100% | 1 |
| 8 | BIOCOMPU GRP | 49249 | 0% | 100% | 1 |
| 9 | BIOINFORMACT UNIT | 49249 | 0% | 100% | 1 |
| 10 | BIOINFORMAT GRP COMP SCI | 49249 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 14911 | 7% | 1% | 44 |
| 2 | BIOINFORMATICS | 13952 | 9% | 1% | 55 |
| 3 | JOURNAL OF STATISTICAL MECHANICS-THEORY AND EXPERIMENT | 6660 | 4% | 1% | 22 |
| 4 | BMC STRUCTURAL BIOLOGY | 6276 | 1% | 2% | 8 |
| 5 | BMC BIOINFORMATICS | 6269 | 5% | 0% | 31 |
| 6 | PLOS COMPUTATIONAL BIOLOGY | 4669 | 3% | 0% | 21 |
| 7 | PROTEIN ENGINEERING | 3118 | 2% | 1% | 12 |
| 8 | MOLECULAR BIOLOGY AND EVOLUTION | 1965 | 2% | 0% | 15 |
| 9 | NEURAL NETWORKS | 1548 | 2% | 0% | 10 |
| 10 | INTERDISCIPLINARY SCIENCES-COMPUTATIONAL LIFE SCIENCES | 1429 | 0% | 1% | 3 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CORRELATED MUTATIONS | 395345 | 3% | 47% | 17 | Search CORRELATED+MUTATIONS | Search CORRELATED+MUTATIONS |
| 2 | DIRECT COUPLING ANALYSIS | 253281 | 1% | 86% | 6 | Search DIRECT+COUPLING+ANALYSIS | Search DIRECT+COUPLING+ANALYSIS |
| 3 | INVERSE ISING PROBLEM | 246247 | 1% | 100% | 5 | Search INVERSE+ISING+PROBLEM | Search INVERSE+ISING+PROBLEM |
| 4 | CONTACT PREDICTION | 246237 | 2% | 50% | 10 | Search CONTACT+PREDICTION | Search CONTACT+PREDICTION |
| 5 | CONTACT MAP | 222209 | 3% | 24% | 19 | Search CONTACT+MAP | Search CONTACT+MAP |
| 6 | CONTACT MAP PREDICTION | 175888 | 1% | 71% | 5 | Search CONTACT+MAP+PREDICTION | Search CONTACT+MAP+PREDICTION |
| 7 | PROTEIN CONTACT MAP | 157596 | 1% | 80% | 4 | Search PROTEIN+CONTACT+MAP | Search PROTEIN+CONTACT+MAP |
| 8 | RESIDUE RESIDUE CONTACT | 157596 | 1% | 80% | 4 | Search RESIDUE+RESIDUE+CONTACT | Search RESIDUE+RESIDUE+CONTACT |
| 9 | COEVOLVING RESIDUES | 147748 | 0% | 100% | 3 | Search COEVOLVING+RESIDUES | Search COEVOLVING+RESIDUES |
| 10 | CORRELATED SUBSTITUTIONS | 147748 | 0% | 100% | 3 | Search CORRELATED+SUBSTITUTIONS | Search CORRELATED+SUBSTITUTIONS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | XU, HS , LI, XQ , ZHANG, ZD , SONG, JN , (2013) IDENTIFYING COEVOLUTION BETWEEN AMINO ACID RESIDUES IN PROTEIN FAMILIES: ADVANCES IN THE IMPROVEMENT AND EVALUATION OF CORRELATED MUTATION ALGORITHMS.CURRENT BIOINFORMATICS. VOL. 8. ISSUE 2. P. 148-160 | 54 | 71% | 1 |
| 2 | EKEBERG, M , LOVKVIST, C , LAN, YH , WEIGT, M , AURELL, E , (2013) IMPROVED CONTACT PREDICTION IN PROTEINS: USING PSEUDOLIKELIHOODS TO INFER POTTS MODELS.PHYSICAL REVIEW E. VOL. 87. ISSUE 1. P. - | 29 | 78% | 80 |
| 3 | EKEBERG, M , HARTONEN, T , AURELL, E , (2014) FAST PSEUDOLIKELIHOOD MAXIMIZATION FOR DIRECT-COUPLING ANALYSIS OF PROTEIN STRUCTURE FROM MANY HOMOLOGOUS AMINO-ACID SEQUENCES.JOURNAL OF COMPUTATIONAL PHYSICS. VOL. 276. ISSUE . P. 341 -356 | 30 | 83% | 7 |
| 4 | DE JUAN, D , PAZOS, F , VALENCIA, A , (2013) EMERGING METHODS IN PROTEIN CO-EVOLUTION.NATURE REVIEWS GENETICS. VOL. 14. ISSUE 4. P. 249 -261 | 40 | 39% | 131 |
| 5 | ASHKENAZY, H , KLIGER, Y , (2010) REDUCING PHYLOGENETIC BIAS IN CORRELATED MUTATION ANALYSIS.PROTEIN ENGINEERING DESIGN & SELECTION. VOL. 23. ISSUE 5. P. 321 -326 | 39 | 67% | 13 |
| 6 | FEINAUER, C , SKWARK, MJ , PAGNANI, A , AURELL, E , (2014) IMPROVING CONTACT PREDICTION ALONG THREE DIMENSIONS.PLOS COMPUTATIONAL BIOLOGY. VOL. 10. ISSUE 10. P. - | 28 | 80% | 8 |
| 7 | BARTON, JP , DE LEONARDIS, E , COUCKE, A , COCCO, S , (2016) ACE: ADAPTIVE CLUSTER EXPANSION FOR MAXIMUM ENTROPY GRAPHICAL MODEL INFERENCE.BIOINFORMATICS. VOL. 32. ISSUE 20. P. 3089 -3097 | 27 | 73% | 2 |
| 8 | YANG, J , JIN, QY , ZHANG, B , SHEN, HB , (2016) R2C: IMPROVING AB INITIO RESIDUE CONTACT MAP PREDICTION USING DYNAMIC FUSION STRATEGY AND GAUSSIAN NOISE FILTER.BIOINFORMATICS. VOL. 32. ISSUE 16. P. 2435 -2443 | 29 | 76% | 0 |
| 9 | JEONG, CS , KIM, D , (2012) RELIABLE AND ROBUST DETECTION OF COEVOLVING PROTEIN RESIDUES.PROTEIN ENGINEERING DESIGN & SELECTION. VOL. 25. ISSUE 11. P. 705-713 | 32 | 71% | 11 |
| 10 | PELE, J , MOREAU, M , ABDI, H , RODIEN, P , CASTEL, H , CHABBERT, M , (2014) COMPARATIVE ANAYSIS OF SEQUENCE CONVARIATION METHODS TO MINE EVOLUTIONARY HUBS: EXAMPLES FROM SELECTED GPCR FAMILIES..PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 82. ISSUE 9. P. 2141 -2156 | 36 | 61% | 4 |
Classes with closest relation at Level 1 |