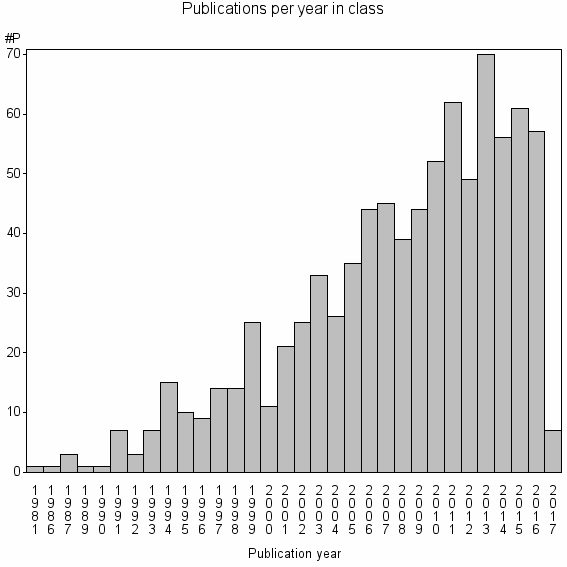
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 13291 | 848 | 46.5 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 13291 | 1 | COMPUTATIONAL PROTEIN DESIGN//PROTEIN DESIGN//DEAD END ELIMINATION | 848 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | COMPUTATIONAL PROTEIN DESIGN | authKW | 2186105 | 10% | 71% | 85 |
| 2 | PROTEIN DESIGN | authKW | 733650 | 16% | 15% | 134 |
| 3 | DEAD END ELIMINATION | authKW | 657846 | 3% | 70% | 26 |
| 4 | COMPUTATIONAL ENZYME DESIGN | authKW | 542225 | 2% | 94% | 16 |
| 5 | ROTAMER LIBRARY | authKW | 364557 | 2% | 56% | 18 |
| 6 | SIDE CHAIN PREDICTION | authKW | 276590 | 2% | 59% | 13 |
| 7 | ENZYME DESIGN | authKW | 220096 | 2% | 34% | 18 |
| 8 | ACTIVE SITE RECAPITULATION | authKW | 180036 | 1% | 100% | 5 |
| 9 | SIDE CHAIN PLACEMENT | authKW | 177260 | 1% | 62% | 8 |
| 10 | DE NOVO PROTEIN DESIGN | authKW | 172114 | 2% | 34% | 14 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 5656 | 66% | 0% | 561 |
| 2 | Biophysics | 2695 | 21% | 0% | 178 |
| 3 | Mathematical & Computational Biology | 1169 | 7% | 0% | 60 |
| 4 | Biochemical Research Methods | 774 | 11% | 0% | 92 |
| 5 | Biotechnology & Applied Microbiology | 247 | 9% | 0% | 73 |
| 6 | Chemistry, Multidisciplinary | 210 | 13% | 0% | 110 |
| 7 | Computer Science, Interdisciplinary Applications | 152 | 4% | 0% | 36 |
| 8 | Cell Biology | 89 | 7% | 0% | 62 |
| 9 | Statistics & Probability | 21 | 2% | 0% | 15 |
| 10 | Physics, Atomic, Molecular & Chemical | 5 | 2% | 0% | 20 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | INTERDISCIPLINARY PROGRAM BIOMOL STRUCT DESIGN | 81015 | 0% | 75% | 3 |
| 2 | BIOCHEM MOL BIOPHYS OPT | 46292 | 0% | 43% | 3 |
| 3 | CHANDLEE | 46292 | 0% | 43% | 3 |
| 4 | PROGRAM BIOL PHYS STRUCT DESIGN | 38402 | 0% | 27% | 4 |
| 5 | ADV MATH ENERGY PLICAT | 36007 | 0% | 100% | 1 |
| 6 | BIOCHEM BIOMOL STRUCT DESIGN BMSD | 36007 | 0% | 100% | 1 |
| 7 | BIOCHEM OPT BIOL | 36007 | 0% | 100% | 1 |
| 8 | BIOCHEM OPT CHEM | 36007 | 0% | 100% | 1 |
| 9 | BIOCHEM OPT CHEM ENGN | 36007 | 0% | 100% | 1 |
| 10 | BIOL SYST SYNTH | 36007 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEIN SCIENCE | 33137 | 9% | 1% | 76 |
| 2 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 31713 | 9% | 1% | 75 |
| 3 | PROTEIN ENGINEERING DESIGN & SELECTION | 13836 | 2% | 2% | 20 |
| 4 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 12403 | 5% | 1% | 46 |
| 5 | CURRENT OPINION IN STRUCTURAL BIOLOGY | 9856 | 3% | 1% | 25 |
| 6 | JOURNAL OF MOLECULAR BIOLOGY | 9111 | 9% | 0% | 77 |
| 7 | CURRENT OPINION IN CHEMICAL BIOLOGY | 4287 | 2% | 1% | 15 |
| 8 | PROTEIN ENGINEERING | 3099 | 2% | 1% | 14 |
| 9 | STRUCTURE | 2716 | 2% | 0% | 17 |
| 10 | JOURNAL OF COMPUTATIONAL BIOLOGY | 2293 | 1% | 1% | 10 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | COMPUTATIONAL PROTEIN DESIGN | 2186105 | 10% | 71% | 85 | Search COMPUTATIONAL+PROTEIN+DESIGN | Search COMPUTATIONAL+PROTEIN+DESIGN |
| 2 | PROTEIN DESIGN | 733650 | 16% | 15% | 134 | Search PROTEIN+DESIGN | Search PROTEIN+DESIGN |
| 3 | DEAD END ELIMINATION | 657846 | 3% | 70% | 26 | Search DEAD+END+ELIMINATION | Search DEAD+END+ELIMINATION |
| 4 | COMPUTATIONAL ENZYME DESIGN | 542225 | 2% | 94% | 16 | Search COMPUTATIONAL+ENZYME+DESIGN | Search COMPUTATIONAL+ENZYME+DESIGN |
| 5 | ROTAMER LIBRARY | 364557 | 2% | 56% | 18 | Search ROTAMER+LIBRARY | Search ROTAMER+LIBRARY |
| 6 | SIDE CHAIN PREDICTION | 276590 | 2% | 59% | 13 | Search SIDE+CHAIN+PREDICTION | Search SIDE+CHAIN+PREDICTION |
| 7 | ENZYME DESIGN | 220096 | 2% | 34% | 18 | Search ENZYME+DESIGN | Search ENZYME+DESIGN |
| 8 | ACTIVE SITE RECAPITULATION | 180036 | 1% | 100% | 5 | Search ACTIVE+SITE+RECAPITULATION | Search ACTIVE+SITE+RECAPITULATION |
| 9 | SIDE CHAIN PLACEMENT | 177260 | 1% | 62% | 8 | Search SIDE+CHAIN+PLACEMENT | Search SIDE+CHAIN+PLACEMENT |
| 10 | DE NOVO PROTEIN DESIGN | 172114 | 2% | 34% | 14 | Search DE+NOVO+PROTEIN+DESIGN | Search DE+NOVO+PROTEIN+DESIGN |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MANDELL, DJ , KORTEMME, T , (2009) COMPUTER-AIDED DESIGN OF FUNCTIONAL PROTEIN INTERACTIONS.NATURE CHEMICAL BIOLOGY. VOL. 5. ISSUE 11. P. 797 -807 | 72 | 75% | 91 |
| 2 | LONDON, N , AMBROGGIO, X , (2014) AN ACCURATE BINDING INTERACTION MODEL IN DE NOVO COMPUTATIONAL PROTEIN DESIGN OF INTERACTIONS: IF YOU BUILD IT, THEY WILL BIND.JOURNAL OF STRUCTURAL BIOLOGY. VOL. 185. ISSUE 2. P. 136-146 | 67 | 66% | 8 |
| 3 | SAMISH, I , MACDERMAID, CM , PEREZ-AGUILAR, JM , SAVEN, JG , (2011) THEORETICAL AND COMPUTATIONAL PROTEIN DESIGN.ANNUAL REVIEW OF PHYSICAL CHEMISTRY, VOL 62. VOL. 62. ISSUE . P. 129 -149 | 77 | 54% | 62 |
| 4 | CANUTESCU, AA , SHELENKOV, AA , DUNBRACK, RL , (2003) A GRAPH-THEORY ALGORITHM FOR RAPID PROTEIN SIDE-CHAIN PREDICTION.PROTEIN SCIENCE. VOL. 12. ISSUE 9. P. 2001-2014 | 37 | 90% | 684 |
| 5 | GAILLARD, T , SIMONSON, T , (2014) PAIRWISE DECOMPOSITION OF AN MMGBSA ENERGY FUNCTION FOR COMPUTATIONAL PROTEIN DESIGN.JOURNAL OF COMPUTATIONAL CHEMISTRY. VOL. 35. ISSUE 18. P. 1371 -1387 | 66 | 65% | 5 |
| 6 | GAILLARD, T , PANEL, N , SIMONSON, T , (2016) PROTEIN SIDE CHAIN CONFORMATION PREDICTIONS WITH AN MMGBSA ENERGY FUNCTION.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 84. ISSUE 6. P. 803 -819 | 70 | 58% | 0 |
| 7 | HUANG, YM , BYSTROFF, C , (2013) EXPANDED EXPLORATIONS INTO THE OPTIMIZATION OF AN ENERGY FUNCTION FOR PROTEIN DESIGN.IEEE-ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS. VOL. 10. ISSUE 5. P. 1176-1187 | 49 | 83% | 1 |
| 8 | LASSILA, JK , (2010) CONFORMATIONAL DIVERSITY AND COMPUTATIONAL ENZYME DESIGN.CURRENT OPINION IN CHEMICAL BIOLOGY. VOL. 14. ISSUE 5. P. 676-682 | 49 | 79% | 16 |
| 9 | FRANCIS-LYON, P , KOEHL, P , (2014) PROTEIN SIDE-CHAIN MODELING WITH A PROTEIN-DEPENDENT OPTIMIZED ROTAMER LIBRARY.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 82. ISSUE 9. P. 2000 -2017 | 47 | 73% | 5 |
| 10 | GAINZA, P , NISONOFF, HM , DONALD, BR , (2016) ALGORITHMS FOR PROTEIN DESIGN.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 39. ISSUE . P. 16 -26 | 49 | 65% | 1 |
Classes with closest relation at Level 1 |