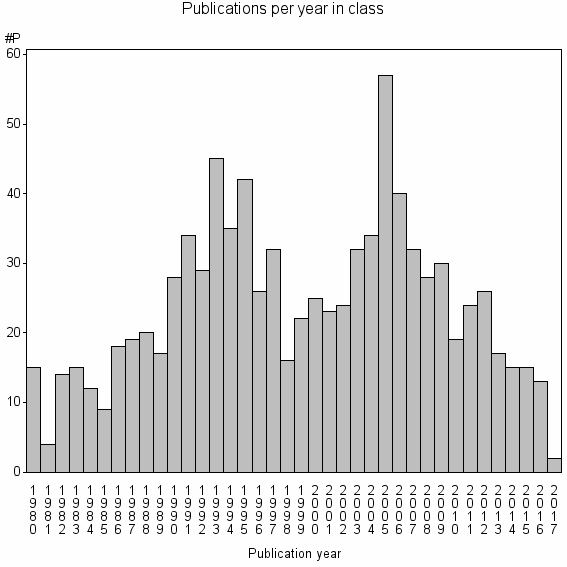
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 12405 | 908 | 37.0 | 65% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN SECONDARY STRUCTURE PREDICTION | authKW | 896714 | 4% | 67% | 40 |
| 2 | SECONDARY STRUCTURE PREDICTION | authKW | 760830 | 8% | 31% | 74 |
| 3 | SOLVENT ACCESSIBILITY | authKW | 279616 | 4% | 24% | 34 |
| 4 | COMPOUND PYRAMID MODEL | authKW | 235394 | 1% | 100% | 7 |
| 5 | PROTEIN STRUCTURE PREDICTION | authKW | 194412 | 8% | 8% | 70 |
| 6 | CHAMELEON SEQUENCES | authKW | 134507 | 1% | 67% | 6 |
| 7 | SOLVENT ACCESSIBILITY PREDICTION | authKW | 120096 | 1% | 71% | 5 |
| 8 | BETA TURN PREDICTION | authKW | 107607 | 0% | 80% | 4 |
| 9 | RESIDUE DEPTH | authKW | 107607 | 0% | 80% | 4 |
| 10 | PREDICTION CONTEST | authKW | 100883 | 0% | 100% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 5216 | 14% | 0% | 128 |
| 2 | Biochemistry & Molecular Biology | 4008 | 55% | 0% | 498 |
| 3 | Biophysics | 3029 | 21% | 0% | 195 |
| 4 | Biochemical Research Methods | 2833 | 19% | 0% | 174 |
| 5 | Computer Science, Interdisciplinary Applications | 1410 | 11% | 0% | 101 |
| 6 | Biotechnology & Applied Microbiology | 514 | 11% | 0% | 103 |
| 7 | Computer Science, Artificial Intelligence | 402 | 6% | 0% | 55 |
| 8 | Biology | 292 | 6% | 0% | 52 |
| 9 | Computer Science, Theory & Methods | 82 | 3% | 0% | 30 |
| 10 | Medical Informatics | 68 | 1% | 0% | 11 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIO BIOPHYS CHEM ABT KLINGELBERGSTR 70 | 44836 | 0% | 67% | 2 |
| 2 | ANALYT BIOSTAT SECT12 S MSC 5626 | 33628 | 0% | 100% | 1 |
| 3 | BIOHYS MOL BIOL | 33628 | 0% | 100% | 1 |
| 4 | BIOL CHIM PROTBIOCH STRUCT FONCT | 33628 | 0% | 100% | 1 |
| 5 | BIOMED TECHNOL HUMAN GENET | 33628 | 0% | 100% | 1 |
| 6 | BIOTECHNOL GENOMIC SCI PROGRAM | 33628 | 0% | 100% | 1 |
| 7 | BIOTECNOL MOLGRP BIOTECNOL AMBIENTAL IND | 33628 | 0% | 100% | 1 |
| 8 | CHEM L PASTEURA 1 | 33628 | 0% | 100% | 1 |
| 9 | CHRISTOPHER S BOND LIFE SCI 271C | 33628 | 0% | 100% | 1 |
| 10 | CNRS UMR 5086 BIOINFORMAT RMN STRUCT | 33628 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 50574 | 4% | 4% | 37 |
| 2 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 32858 | 9% | 1% | 79 |
| 3 | PROTEIN ENGINEERING | 31437 | 5% | 2% | 46 |
| 4 | PROTEINS-STRUCTURE FUNCTION AND GENETICS | 9143 | 1% | 2% | 11 |
| 5 | BIOINFORMATICS | 6625 | 5% | 0% | 46 |
| 6 | INTERNATIONAL JOURNAL OF PEPTIDE AND PROTEIN RESEARCH | 5890 | 2% | 1% | 21 |
| 7 | PROTEIN SCIENCE | 4784 | 3% | 0% | 30 |
| 8 | JOURNAL OF MOLECULAR GRAPHICS | 3812 | 1% | 2% | 5 |
| 9 | BMC BIOINFORMATICS | 3725 | 3% | 0% | 29 |
| 10 | COMPUTATIONAL BIOLOGY AND CHEMISTRY | 2444 | 1% | 1% | 8 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | DOR, O , ZHOU, YQ , (2007) ACHIEVING 80% TEN-FOLD CROSS-VALIDATED ACCURACY FOR SECONDARY STRUCTURE PREDICTION BY LARGE-SCALE TRAINING.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 66. ISSUE 4. P. 838 -845 | 43 | 93% | 44 |
| 2 | SIVAN, S , FILO, O , SIEGELMANN, H , (2007) APPLICATION OF EXPERT NETWORKS FOR PREDICTING PROTEINS SECONDARY STRUCTURE.BIOMOLECULAR ENGINEERING. VOL. 24. ISSUE 2. P. 237-243 | 47 | 84% | 4 |
| 3 | KURGAN, L , DISFANI, FM , (2011) STRUCTURAL PROTEIN DESCRIPTORS IN 1-DIMENSION AND THEIR SEQUENCE-BASED PREDICTIONS.CURRENT PROTEIN & PEPTIDE SCIENCE. VOL. 12. ISSUE 6. P. 470 -489 | 65 | 41% | 5 |
| 4 | ZANGOOEI, MH , JALILI, S , (2012) PROTEIN SECONDARY STRUCTURE PREDICTION USING DWKF BASED ON SVR-NSGAII.NEUROCOMPUTING. VOL. 94. ISSUE . P. 87 -101 | 34 | 76% | 7 |
| 5 | ROST, B , SANDER, C , (1993) PREDICTION OF PROTEIN SECONDARY STRUCTURE AT BETTER THAN 70-PERCENT ACCURACY.JOURNAL OF MOLECULAR BIOLOGY. VOL. 232. ISSUE 2. P. 584 -599 | 28 | 56% | 2325 |
| 6 | GHANTY, P , PAL, NR , MUDI, RK , (2013) PREDICTION OF PROTEIN SECONDARY STRUCTURE USING PROBABILITY BASED FEATURES AND A HYBRID SYSTEM.JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY. VOL. 11. ISSUE 5. P. - | 32 | 78% | 0 |
| 7 | ZANGOOEI, MH , JALILI, S , (2012) PSSP WITH DYNAMIC WEIGHTED KERNEL FUSION BASED ON SVM-PHGS.KNOWLEDGE-BASED SYSTEMS. VOL. 27. ISSUE . P. 424 -442 | 32 | 74% | 4 |
| 8 | SARASWATHI, S , FERNANDEZ-MARTINEZ, JL , KOLINSKI, A , JERNIGAN, RL , KLOCZKOWSKI, A , (2013) DISTRIBUTIONS OF AMINO ACIDS SUGGEST THAT CERTAIN RESIDUE TYPES MORE EFFECTIVELY DETERMINE PROTEIN SECONDARY STRUCTURE.JOURNAL OF MOLECULAR MODELING. VOL. 19. ISSUE 10. P. 4337 -4348 | 31 | 70% | 0 |
| 9 | ROST, B , SANDER, C , (1994) COMBINING EVOLUTIONARY INFORMATION AND NEURAL NETWORKS TO PREDICT PROTEIN SECONDARY STRUCTURE.PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS. VOL. 19. ISSUE 1. P. 55 -72 | 39 | 41% | 1228 |
| 10 | CHENG, HT , SEN, TZ , JERNIGAN, RL , KLOCZKOWSKI, A , (2010) DATA MINING FOR PROTEIN SECONDARY STRUCTURE PREDICTION.DATA MINING IN CRYSTALLOGRAPHY. VOL. 134. ISSUE . P. 135 -167 | 38 | 58% | 0 |
Classes with closest relation at Level 1 |