Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
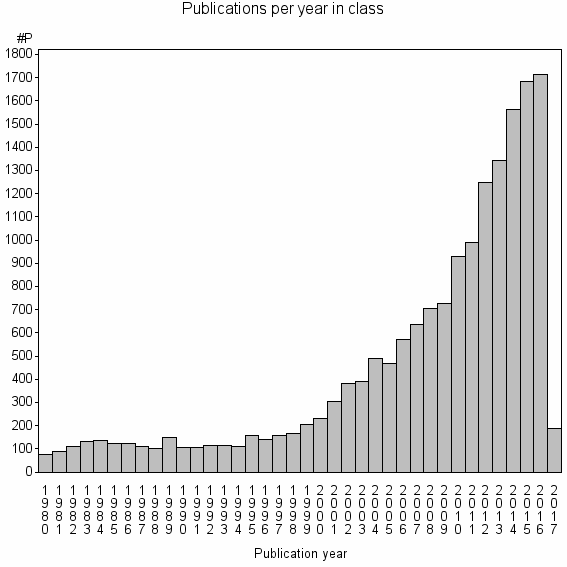
| 382 | 17086 | 46.8 | 87% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA METHYLATION | authKW | 1624241 | 17% | 31% | 2965 |
| 2 | EPIGENETICS | authKW | 450412 | 10% | 15% | 1655 |
| 3 | METHYLATION | authKW | 379622 | 7% | 18% | 1171 |
| 4 | 5 HYDROXYMETHYLCYTOSINE | authKW | 214795 | 1% | 81% | 148 |
| 5 | RASSF1A | authKW | 208858 | 1% | 69% | 169 |
| 6 | CPG ISLAND | authKW | 188974 | 2% | 38% | 279 |
| 7 | CELL FREE DNA | authKW | 171757 | 1% | 54% | 178 |
| 8 | DNA METHYLTRANSFERASE | authKW | 170458 | 2% | 36% | 265 |
| 9 | HYPERMETHYLATION | authKW | 149393 | 1% | 36% | 234 |
| 10 | CIRCULATING DNA | authKW | 149061 | 1% | 80% | 105 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Oncology | 53910 | 30% | 1% | 5131 |
| 2 | Genetics & Heredity | 28231 | 18% | 1% | 3053 |
| 3 | Biochemistry & Molecular Biology | 13181 | 26% | 0% | 4466 |
| 4 | Cell Biology | 10694 | 14% | 0% | 2472 |
| 5 | Pathology | 3356 | 5% | 0% | 792 |
| 6 | Medical Laboratory Technology | 2474 | 2% | 0% | 384 |
| 7 | Biotechnology & Applied Microbiology | 2413 | 6% | 0% | 1110 |
| 8 | Gastroenterology & Hepatology | 1948 | 4% | 0% | 689 |
| 9 | Medicine, Research & Experimental | 1105 | 4% | 0% | 719 |
| 10 | Biochemical Research Methods | 993 | 4% | 0% | 602 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EPIGENET | 106615 | 1% | 27% | 220 |
| 2 | CANC EPIGENET | 52973 | 1% | 26% | 116 |
| 3 | EXCELLENCE MOL GENET CANC HUMAN DIS | 37898 | 0% | 59% | 36 |
| 4 | EPIGEN | 28526 | 0% | 30% | 54 |
| 5 | CONJOINT GASTROENTEROL | 26258 | 0% | 39% | 38 |
| 6 | ENVIRONM EPIGENET | 25927 | 0% | 58% | 25 |
| 7 | EPIGENET PROGRAMME | 23409 | 0% | 39% | 34 |
| 8 | STAT GENOM GRP | 22235 | 0% | 69% | 18 |
| 9 | EPIGENET GRP | 20971 | 0% | 37% | 32 |
| 10 | PATHOL | 20944 | 14% | 1% | 2401 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CLINICAL EPIGENETICS | 125386 | 1% | 42% | 169 |
| 2 | EPIGENOMICS | 108845 | 1% | 36% | 168 |
| 3 | EPIGENETICS | 56368 | 1% | 35% | 90 |
| 4 | CIBA FOUNDATION SYMPOSIA | 44321 | 2% | 10% | 261 |
| 5 | EPIGENETICS & CHROMATIN | 16618 | 0% | 18% | 52 |
| 6 | CLINICAL CANCER RESEARCH | 8389 | 2% | 2% | 271 |
| 7 | GENOME BIOLOGY | 8125 | 1% | 4% | 118 |
| 8 | GENOME RESEARCH | 5879 | 0% | 5% | 68 |
| 9 | CANCER RESEARCH | 5756 | 2% | 1% | 373 |
| 10 | NUCLEIC ACIDS RESEARCH | 5444 | 2% | 1% | 366 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA METHYLATION | 1624241 | 17% | 31% | 2965 | Search DNA+METHYLATION | Search DNA+METHYLATION |
| 2 | EPIGENETICS | 450412 | 10% | 15% | 1655 | Search EPIGENETICS | Search EPIGENETICS |
| 3 | METHYLATION | 379622 | 7% | 18% | 1171 | Search METHYLATION | Search METHYLATION |
| 4 | 5 HYDROXYMETHYLCYTOSINE | 214795 | 1% | 81% | 148 | Search 5+HYDROXYMETHYLCYTOSINE | Search 5+HYDROXYMETHYLCYTOSINE |
| 5 | RASSF1A | 208858 | 1% | 69% | 169 | Search RASSF1A | Search RASSF1A |
| 6 | CPG ISLAND | 188974 | 2% | 38% | 279 | Search CPG+ISLAND | Search CPG+ISLAND |
| 7 | CELL FREE DNA | 171757 | 1% | 54% | 178 | Search CELL+FREE+DNA | Search CELL+FREE+DNA |
| 8 | DNA METHYLTRANSFERASE | 170458 | 2% | 36% | 265 | Search DNA+METHYLTRANSFERASE | Search DNA+METHYLTRANSFERASE |
| 9 | HYPERMETHYLATION | 149393 | 1% | 36% | 234 | Search HYPERMETHYLATION | Search HYPERMETHYLATION |
| 10 | CIRCULATING DNA | 149061 | 1% | 80% | 105 | Search CIRCULATING+DNA | Search CIRCULATING+DNA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WU, H , ZHANG, Y , (2014) REVERSING DNA METHYLATION: MECHANISMS, GENOMICS, AND BIOLOGICAL FUNCTIONS.CELL. VOL. 156. ISSUE 1-2. P. 45-68 | 164 | 78% | 228 |
| 2 | FLEISCHHACKER, M , SCHMIDT, B , (2007) CIRCULATING NUCLEIC ACIDS (CNAS) AND CANCER - A SURVEY.BIOCHIMICA ET BIOPHYSICA ACTA-REVIEWS ON CANCER. VOL. 1775. ISSUE 1. P. 181 -232 | 236 | 64% | 314 |
| 3 | SCHUBELER, D , (2015) FUNCTION AND INFORMATION CONTENT OF DNA METHYLATION.NATURE. VOL. 517. ISSUE 7534. P. 321 -326 | 86 | 74% | 197 |
| 4 | PASTOR, WA , ARAVIND, L , RAO, A , (2013) TETONIC SHIFT: BIOLOGICAL ROLES OF TET PROTEINS IN DNA DEMETHYLATION AND TRANSCRIPTION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 14. ISSUE 6. P. 341 -356 | 130 | 73% | 213 |
| 5 | SMITH, ZD , MEISSNER, A , (2013) DNA METHYLATION: ROLES IN MAMMALIAN DEVELOPMENT.NATURE REVIEWS GENETICS. VOL. 14. ISSUE 3. P. 204 -220 | 114 | 58% | 500 |
| 6 | JUNG, K , FLEISCHHACKER, M , RABIEN, A , (2010) CELL-FREE DNA IN THE BLOOD AS A SOLID TUMOR BIOMARKER-A CRITICAL APPRAISAL OF THE LITERATURE.CLINICA CHIMICA ACTA. VOL. 411. ISSUE 21-22. P. 1611-1624 | 172 | 83% | 113 |
| 7 | RASMUSSEN, KD , HELIN, K , (2016) ROLE OF TET ENZYMES IN DNA METHYLATION, DEVELOPMENT, AND CANCER.GENES & DEVELOPMENT. VOL. 30. ISSUE 7. P. 733 -750 | 109 | 72% | 20 |
| 8 | SCHWARZENBACH, H , HOON, DSB , PANTEL, K , (2011) CELL-FREE NUCLEIC ACIDS AS BIOMARKERS IN CANCER PATIENTS.NATURE REVIEWS CANCER. VOL. 11. ISSUE 6. P. 426 -437 | 110 | 66% | 606 |
| 9 | LAIRD, PW , (2010) PRINCIPLES AND CHALLENGES OF GENOME-WIDE DNA METHYLATION ANALYSIS.NATURE REVIEWS GENETICS. VOL. 11. ISSUE 3. P. 191 -203 | 107 | 69% | 629 |
| 10 | JURKOWSKA, RZ , JURKOWSKI, TP , JELTSCH, A , (2011) STRUCTURE AND FUNCTION OF MAMMALIAN DNA METHYLTRANSFERASES.CHEMBIOCHEM. VOL. 12. ISSUE 2. P. 206 -222 | 142 | 70% | 214 |
Classes with closest relation at Level 2 |