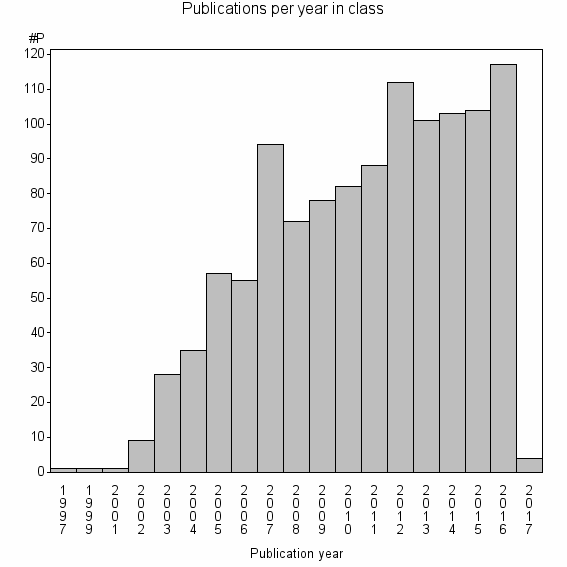
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 9574 | 1142 | 34.6 | 79% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 868 | 2 | BMC BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS | 11572 |
| 9574 | 1 | GENE SET ANALYSIS//GENE ONTOLOGY//BMC BIOINFORMATICS | 1142 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENE SET ANALYSIS | authKW | 415915 | 3% | 47% | 33 |
| 2 | GENE ONTOLOGY | authKW | 263910 | 8% | 11% | 89 |
| 3 | BMC BIOINFORMATICS | journal | 161843 | 19% | 3% | 213 |
| 4 | BIOINFORMATICS | journal | 102575 | 18% | 2% | 202 |
| 5 | MATHEMATICAL & COMPUTATIONAL BIOLOGY | WoSSC | 97213 | 53% | 1% | 609 |
| 6 | ENRICHMENT ANALYSIS | authKW | 91158 | 2% | 19% | 18 |
| 7 | GENE SET | authKW | 85236 | 1% | 25% | 13 |
| 8 | RELATED TERMS | authKW | 80211 | 0% | 100% | 3 |
| 9 | GENE SET ENRICHMENT | authKW | 69848 | 1% | 29% | 9 |
| 10 | SEMANTIC SIMILARITY MEASURE | authKW | 68944 | 1% | 37% | 7 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 97213 | 53% | 1% | 609 |
| 2 | Biochemical Research Methods | 26497 | 50% | 0% | 576 |
| 3 | Biotechnology & Applied Microbiology | 16390 | 50% | 0% | 576 |
| 4 | Statistics & Probability | 733 | 7% | 0% | 76 |
| 5 | Genetics & Heredity | 732 | 12% | 0% | 134 |
| 6 | Medical Informatics | 606 | 3% | 0% | 34 |
| 7 | Biochemistry & Molecular Biology | 475 | 21% | 0% | 236 |
| 8 | Computer Science, Interdisciplinary Applications | 393 | 6% | 0% | 64 |
| 9 | Multidisciplinary Sciences | 164 | 2% | 0% | 26 |
| 10 | Computer Science, Artificial Intelligence | 32 | 2% | 0% | 23 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | FUNCT GENOM NODE | 61863 | 1% | 26% | 9 |
| 2 | BIOINFORMAT FUNCT GENOM NODE INB | 53474 | 0% | 100% | 2 |
| 3 | CIHR MSFHR | 53474 | 0% | 100% | 2 |
| 4 | GOA PROJECT | 53474 | 0% | 100% | 2 |
| 5 | HOLM GRP | 53474 | 0% | 100% | 2 |
| 6 | NIAID OFF TECHNOL INFORMAT SYST | 53474 | 0% | 100% | 2 |
| 7 | BIOINFORMAT RARE DIS BIER | 45826 | 1% | 29% | 6 |
| 8 | INB | 39292 | 1% | 10% | 15 |
| 9 | FUNCT GENOM NODE INB | 37420 | 1% | 20% | 7 |
| 10 | BIOINFORMAT SYST BIOL LIFE SCI | 35648 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC BIOINFORMATICS | 161843 | 19% | 3% | 213 |
| 2 | BIOINFORMATICS | 102575 | 18% | 2% | 202 |
| 3 | BRIEFINGS IN BIOINFORMATICS | 8627 | 1% | 2% | 15 |
| 4 | NUCLEIC ACIDS RESEARCH | 8400 | 10% | 0% | 112 |
| 5 | IEEE-ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS | 6805 | 1% | 2% | 17 |
| 6 | BMC SYSTEMS BIOLOGY | 5987 | 2% | 1% | 19 |
| 7 | STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY | 4271 | 1% | 2% | 9 |
| 8 | GENOME BIOLOGY | 3919 | 2% | 1% | 21 |
| 9 | PLOS COMPUTATIONAL BIOLOGY | 2763 | 2% | 0% | 22 |
| 10 | JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY | 2618 | 1% | 2% | 6 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENE SET ANALYSIS | 415915 | 3% | 47% | 33 | Search GENE+SET+ANALYSIS | Search GENE+SET+ANALYSIS |
| 2 | GENE ONTOLOGY | 263910 | 8% | 11% | 89 | Search GENE+ONTOLOGY | Search GENE+ONTOLOGY |
| 3 | ENRICHMENT ANALYSIS | 91158 | 2% | 19% | 18 | Search ENRICHMENT+ANALYSIS | Search ENRICHMENT+ANALYSIS |
| 4 | GENE SET | 85236 | 1% | 25% | 13 | Search GENE+SET | Search GENE+SET |
| 5 | RELATED TERMS | 80211 | 0% | 100% | 3 | Search RELATED+TERMS | Search RELATED+TERMS |
| 6 | GENE SET ENRICHMENT | 69848 | 1% | 29% | 9 | Search GENE+SET+ENRICHMENT | Search GENE+SET+ENRICHMENT |
| 7 | SEMANTIC SIMILARITY MEASURE | 68944 | 1% | 37% | 7 | Search SEMANTIC+SIMILARITY+MEASURE | Search SEMANTIC+SIMILARITY+MEASURE |
| 8 | SEMANTIC SIMILARITY | 61850 | 2% | 9% | 26 | Search SEMANTIC+SIMILARITY | Search SEMANTIC+SIMILARITY |
| 9 | BIOLOGICAL SEARCH ENGINE | 53474 | 0% | 100% | 2 | Search BIOLOGICAL+SEARCH+ENGINE | Search BIOLOGICAL+SEARCH+ENGINE |
| 10 | BIOMOLECULAR ANNOTATIONS | 53474 | 0% | 100% | 2 | Search BIOMOLECULAR+ANNOTATIONS | Search BIOMOLECULAR+ANNOTATIONS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | HUANG, DW , SHERMAN, BT , LEMPICKI, RA , (2009) SYSTEMATIC AND INTEGRATIVE ANALYSIS OF LARGE GENE LISTS USING DAVID BIOINFORMATICS RESOURCES.NATURE PROTOCOLS. VOL. 4. ISSUE 1. P. 44-57 | 10 | 83% | 8308 |
| 2 | KHATRI, P , SIROTA, M , BUTTE, AJ , (2012) TEN YEARS OF PATHWAY ANALYSIS: CURRENT APPROACHES AND OUTSTANDING CHALLENGES.PLOS COMPUTATIONAL BIOLOGY. VOL. 8. ISSUE 2. P. - | 45 | 58% | 334 |
| 3 | NAM, D , KIM, SY , (2008) GENE-SET APPROACH FOR EXPRESSION PATTERN ANALYSIS.BRIEFINGS IN BIOINFORMATICS. VOL. 9. ISSUE 3. P. 189-197 | 39 | 89% | 169 |
| 4 | DOPAZO, J , (2009) FORMULATING AND TESTING HYPOTHESES IN FUNCTIONAL GENOMICS.ARTIFICIAL INTELLIGENCE IN MEDICINE. VOL. 45. ISSUE 2-3. P. 97-107 | 63 | 58% | 6 |
| 5 | ACKERMANN, M , STRIMMER, K , (2009) A GENERAL MODULAR FRAMEWORK FOR GENE SET ENRICHMENT ANALYSIS.BMC BIOINFORMATICS. VOL. 10. ISSUE . P. - | 29 | 78% | 143 |
| 6 | AL-SHAHROUR, F , CARBONELL, J , MINGUEZ, P , GOETZ, S , CONESA, A , TARRRAGA, J , MEDINA, I , ALLOZA, E , MONTANER, D , DOPAZO, J , (2008) BABELOMICS: ADVANCED FUNCTIONAL PROFILING OF TRANSCRIPTOMICS, PROTEOMICS AND GENOMICS EXPERIMENTS.NUCLEIC ACIDS RESEARCH. VOL. 36. ISSUE . P. W341-W346 | 39 | 71% | 37 |
| 7 | ZHENG, Q , WANG, XJ , (2008) GOEAST: A WEB-BASED SOFTWARE TOOLKIT FOR GENE ONTOLOGY ENRICHMENT ANALYSIS.NUCLEIC ACIDS RESEARCH. VOL. 36. ISSUE . P. W358-W363 | 23 | 74% | 302 |
| 8 | MACIEJEWSKI, H , (2014) GENE SET ANALYSIS METHODS: STATISTICAL MODELS AND METHODOLOGICAL DIFFERENCES.BRIEFINGS IN BIOINFORMATICS. VOL. 15. ISSUE 4. P. 504 -518 | 21 | 91% | 22 |
| 9 | HUANG, DW , SHERMAN, BT , TAN, Q , COLLINS, JR , ALVORD, WG , ROAYAEI, J , STEPHENS, R , BASELER, MW , LANE, HC , LEMPICKI, RA , (2007) THE DAVID GENE FUNCTIONAL CLASSIFICATION TOOL: A NOVEL BIOLOGICAL MODULE-CENTRIC ALGORITHM TO FUNCTIONALLY ANALYZE LARGE GENE LISTS.GENOME BIOLOGY. VOL. 8. ISSUE 9. P. - | 23 | 77% | 299 |
| 10 | GOEMAN, JJ , BUHLMANN, P , (2007) ANALYZING GENE EXPRESSION DATA IN TERMS OF GENE SETS: METHODOLOGICAL ISSUES.BIOINFORMATICS. VOL. 23. ISSUE 8. P. 980 -987 | 21 | 81% | 323 |
Classes with closest relation at Level 1 |