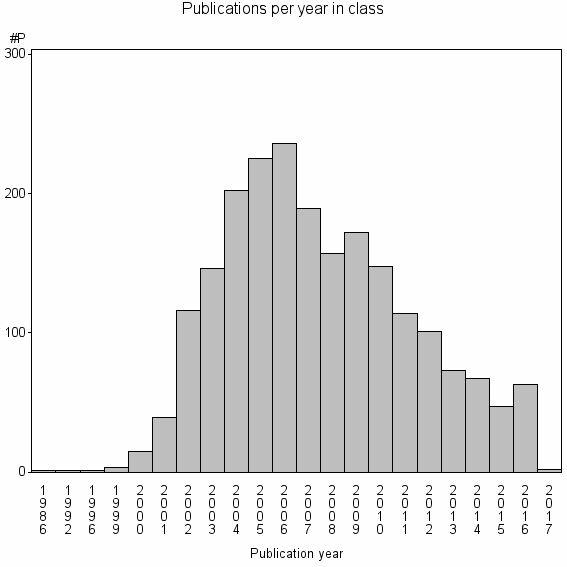
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 2901 | 2118 | 29.9 | 74% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 868 | 2 | BMC BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS | 11572 |
| 2901 | 1 | BMC BIOINFORMATICS//MICROARRAY IMAGE//BIOINFORMATICS | 2118 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | BMC BIOINFORMATICS | journal | 189513 | 15% | 4% | 314 |
| 2 | MICROARRAY IMAGE | authKW | 129738 | 0% | 100% | 9 |
| 3 | BIOINFORMATICS | journal | 107650 | 13% | 3% | 282 |
| 4 | MATHEMATICAL & COMPUTATIONAL BIOLOGY | WoSSC | 105343 | 41% | 1% | 865 |
| 5 | MICROARRAY | authKW | 102377 | 15% | 2% | 325 |
| 6 | MICROARRAY IMAGE PROCESSING | authKW | 86492 | 0% | 100% | 6 |
| 7 | MICROARRAY EXPERIMENTS | authKW | 77835 | 0% | 60% | 9 |
| 8 | DYE BIAS | authKW | 57661 | 0% | 100% | 4 |
| 9 | BATCH EFFECT REMOVAL | authKW | 46127 | 0% | 80% | 4 |
| 10 | MICROARRAY GRIDDING | authKW | 46127 | 0% | 80% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 105343 | 41% | 1% | 865 |
| 2 | Biochemical Research Methods | 30034 | 40% | 0% | 840 |
| 3 | Biotechnology & Applied Microbiology | 23524 | 45% | 0% | 944 |
| 4 | Statistics & Probability | 5323 | 13% | 0% | 267 |
| 5 | Genetics & Heredity | 2726 | 16% | 0% | 338 |
| 6 | Medical Informatics | 875 | 3% | 0% | 56 |
| 7 | Biochemistry & Molecular Biology | 777 | 20% | 0% | 418 |
| 8 | Computer Science, Interdisciplinary Applications | 733 | 6% | 0% | 119 |
| 9 | Biology | 147 | 3% | 0% | 65 |
| 10 | Multidisciplinary Sciences | 115 | 2% | 0% | 32 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ENSEMBL TEAM | 28831 | 0% | 100% | 2 |
| 2 | INFORMAT NANTES ATLANTIQUE LINA | 28831 | 0% | 100% | 2 |
| 3 | MAX MCGEE JUVENILE DIABET | 26410 | 1% | 17% | 11 |
| 4 | IAN POTTER FDN CANC GENOM PREDICT MED | 21620 | 0% | 50% | 3 |
| 5 | BIOMOL ANAL IMAGING | 19219 | 0% | 67% | 2 |
| 6 | TRANSCRIPT GENOM CORE | 19219 | 0% | 67% | 2 |
| 7 | ADV MAT CP NANOSAFETY | 14415 | 0% | 100% | 1 |
| 8 | AI COMO | 14415 | 0% | 100% | 1 |
| 9 | AIRCRAFT SPARE MANAGEMENT | 14415 | 0% | 100% | 1 |
| 10 | ANAT EMBRIOL GENETI ANIM | 14415 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC BIOINFORMATICS | 189513 | 15% | 4% | 314 |
| 2 | BIOINFORMATICS | 107650 | 13% | 3% | 282 |
| 3 | STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY | 45599 | 2% | 8% | 40 |
| 4 | BMC GENOMICS | 14649 | 5% | 1% | 99 |
| 5 | BIOSTATISTICS | 12573 | 1% | 3% | 26 |
| 6 | JOURNAL OF COMPUTATIONAL BIOLOGY | 8837 | 1% | 2% | 31 |
| 7 | OMICS-A JOURNAL OF INTEGRATIVE BIOLOGY | 7387 | 1% | 3% | 19 |
| 8 | COMPARATIVE AND FUNCTIONAL GENOMICS | 6319 | 1% | 3% | 13 |
| 9 | NUCLEIC ACIDS RESEARCH | 6101 | 6% | 0% | 131 |
| 10 | GENOME BIOLOGY | 5863 | 2% | 1% | 35 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MICROARRAY IMAGE | 129738 | 0% | 100% | 9 | Search MICROARRAY+IMAGE | Search MICROARRAY+IMAGE |
| 2 | MICROARRAY | 102377 | 15% | 2% | 325 | Search MICROARRAY | Search MICROARRAY |
| 3 | MICROARRAY IMAGE PROCESSING | 86492 | 0% | 100% | 6 | Search MICROARRAY+IMAGE+PROCESSING | Search MICROARRAY+IMAGE+PROCESSING |
| 4 | MICROARRAY EXPERIMENTS | 77835 | 0% | 60% | 9 | Search MICROARRAY+EXPERIMENTS | Search MICROARRAY+EXPERIMENTS |
| 5 | DYE BIAS | 57661 | 0% | 100% | 4 | Search DYE+BIAS | Search DYE+BIAS |
| 6 | BATCH EFFECT REMOVAL | 46127 | 0% | 80% | 4 | Search BATCH+EFFECT+REMOVAL | Search BATCH+EFFECT+REMOVAL |
| 7 | MICROARRAY GRIDDING | 46127 | 0% | 80% | 4 | Search MICROARRAY+GRIDDING | Search MICROARRAY+GRIDDING |
| 8 | SPOT SEGMENTATION | 46127 | 0% | 80% | 4 | Search SPOT+SEGMENTATION | Search SPOT+SEGMENTATION |
| 9 | CDNA MICROARRAY IMAGES | 43246 | 0% | 100% | 3 | Search CDNA+MICROARRAY+IMAGES | Search CDNA+MICROARRAY+IMAGES |
| 10 | CODELINK | 43246 | 0% | 100% | 3 | Search CODELINK | Search CODELINK |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | YAUK, CL , BERNDT, ML , (2007) REVIEW OF THE LITERATURE EXAMINING THE CORRELATION AMONG DNA MICROARRAY TECHNOLOGIES.ENVIRONMENTAL AND MOLECULAR MUTAGENESIS. VOL. 48. ISSUE 5. P. 380 -394 | 59 | 72% | 37 |
| 2 | ARCHER, KJ , DUMUR, CI , TAYLOR, GS , CHAPLIN, MD , GUISEPPI-ELIE, A , GRANT, G , FERREIRA-GONZALEZ, A , GARRETT, CT , (2007) APPLICATION OF A CORRELATION CORRECTION FACTOR IN A MICROARRAY CROSS-PLATFORM REPRODUCIBILITY STUDY.BMC BIOINFORMATICS. VOL. 8. ISSUE . P. - | 40 | 93% | 3 |
| 3 | LI, WT , (2012) VOLCANO PLOTS IN ANALYZING DIFFERENTIAL EXPRESSIONS WITH MRNA MICROARRAYS.JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY. VOL. 10. ISSUE 6. P. - | 56 | 53% | 21 |
| 4 | LAZAR, C , MEGANCK, S , TAMINAU, J , STEENHOFF, D , COLETTA, A , MOLTER, C , WEISS-SOLIS, DY , DUQUE, R , BERSINI, H , NOWE, A , (2013) BATCH EFFECT REMOVAL METHODS FOR MICROARRAY GENE EXPRESSION DATA INTEGRATION: A SURVEY.BRIEFINGS IN BIOINFORMATICS. VOL. 14. ISSUE 4. P. 469-490 | 36 | 73% | 32 |
| 5 | MURIE, C , WOODY, O , LEE, AY , NADON, R , (2009) COMPARISON OF SMALL N STATISTICAL TESTS OF DIFFERENTIAL EXPRESSION APPLIED TO MICROARRAYS.BMC BIOINFORMATICS. VOL. 10. ISSUE . P. - | 35 | 90% | 32 |
| 6 | SHI, LM , REID, LH , JONES, WD , SHIPPY, R , WARRINGTON, JA , BAKER, SC , COLLINS, PJ , DE LONGUEVILLE, F , KAWASAKI, ES , LEE, KY , ET AL (2006) THE MICROARRAY QUALITY CONTROL (MAQC) PROJECT SHOWS INTER- AND INTRAPLATFORM REPRODUCIBILITY OF GENE EXPRESSION MEASUREMENTS.NATURE BIOTECHNOLOGY. VOL. 24. ISSUE 9. P. 1151 -1161 | 22 | 73% | 995 |
| 7 | TSENG, GC , GHOSH, D , FEINGOLD, E , (2012) COMPREHENSIVE LITERATURE REVIEW AND STATISTICAL CONSIDERATIONS FOR MICROARRAY META-ANALYSIS.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE 9. P. 3785 -3799 | 52 | 44% | 82 |
| 8 | KARAKACH, TK , FLIGHT, RM , DOUGLAS, SE , WENTZELL, PD , (2010) AN INTRODUCTION TO DNA MICROARRAYS FOR GENE EXPRESSION ANALYSIS.CHEMOMETRICS AND INTELLIGENT LABORATORY SYSTEMS. VOL. 104. ISSUE 1. P. 28 -52 | 56 | 50% | 14 |
| 9 | HOLLOWAY, AJ , OSHLACK, A , DIYAGAMA, DS , BOWTELL, DDL , SMYTH, GK , (2006) STATISTICAL ANALYSIS OF AN RNA TITRATION SERIES EVALUATES MICROARRAY PRECISION AND SENSITIVITY ON A WHOLE-ARRAY BASIS.BMC BIOINFORMATICS. VOL. 7. ISSUE . P. - | 40 | 78% | 9 |
| 10 | KUO, WP , LIU, F , TRIMARCHI, J , PUNZO, C , LOMBARDI, M , SARANG, J , WHIPPLE, ME , MAYSURIA, M , SERIKAWA, K , LEE, SY , ET AL (2006) A SEQUENCE-ORIENTED COMPARISON OF GENE EXPRESSION MEASUREMENTS ACROSS DIFFERENT HYBRIDIZATION-BASED TECHNOLOGIES.NATURE BIOTECHNOLOGY. VOL. 24. ISSUE 7. P. 832 -840 | 35 | 76% | 100 |
Classes with closest relation at Level 1 |