Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
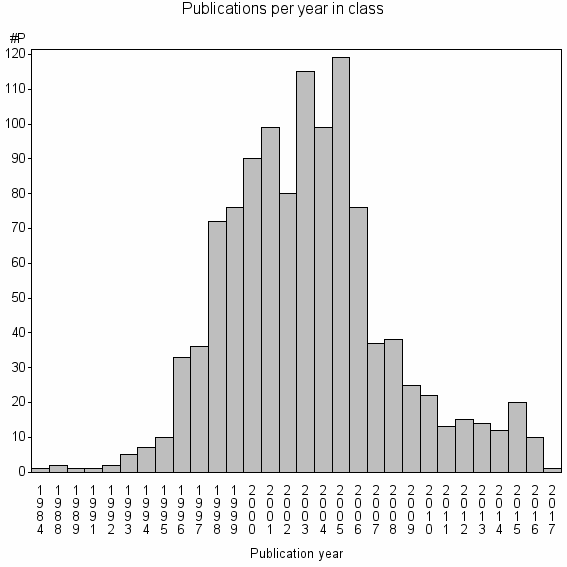
| 9683 | 1131 | 26.7 | 73% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 9683 | 1 | EMBL OUTSTN//LECTURE NOTES IN BIOINFORMATICS//PROT INFORMAT OURCE | 1131 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | EMBL OUTSTN | address | 329634 | 5% | 23% | 53 |
| 2 | LECTURE NOTES IN BIOINFORMATICS | journal | 124919 | 2% | 24% | 19 |
| 3 | PROT INFORMAT OURCE | address | 104121 | 1% | 43% | 9 |
| 4 | BIOINFORMATICS | journal | 88737 | 17% | 2% | 187 |
| 5 | J AN INT PROT INFORMAT DATABASE | address | 80991 | 0% | 100% | 3 |
| 6 | SIMILARITY SCORE DISSECTION | authKW | 80991 | 0% | 100% | 3 |
| 7 | EUROPEAN BIOINFORMAT | address | 71941 | 2% | 10% | 28 |
| 8 | INTERPRO | authKW | 61704 | 0% | 57% | 4 |
| 9 | HIGHER TECH COMP SCI ENGN | address | 60742 | 0% | 75% | 3 |
| 10 | HLTH SCI TECHNOL ICIR | address | 60742 | 0% | 75% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 26130 | 28% | 0% | 316 |
| 2 | Biochemical Research Methods | 8352 | 29% | 0% | 327 |
| 3 | Biotechnology & Applied Microbiology | 6283 | 32% | 0% | 362 |
| 4 | Biochemistry & Molecular Biology | 2485 | 40% | 0% | 456 |
| 5 | Computer Science, Information Systems | 621 | 7% | 0% | 75 |
| 6 | Genetics & Heredity | 562 | 11% | 0% | 119 |
| 7 | Computer Science, Interdisciplinary Applications | 356 | 5% | 0% | 61 |
| 8 | Computer Science, Theory & Methods | 221 | 5% | 0% | 51 |
| 9 | Medical Informatics | 219 | 2% | 0% | 21 |
| 10 | Computer Science, Artificial Intelligence | 81 | 3% | 0% | 32 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EMBL OUTSTN | 329634 | 5% | 23% | 53 |
| 2 | PROT INFORMAT OURCE | 104121 | 1% | 43% | 9 |
| 3 | J AN INT PROT INFORMAT DATABASE | 80991 | 0% | 100% | 3 |
| 4 | EUROPEAN BIOINFORMAT | 71941 | 2% | 10% | 28 |
| 5 | HIGHER TECH COMP SCI ENGN | 60742 | 0% | 75% | 3 |
| 6 | HLTH SCI TECHNOL ICIR | 60742 | 0% | 75% | 3 |
| 7 | BIOMED KNOWLEDGE DISCOVERY TEAMTSURUMI KU | 53994 | 0% | 100% | 2 |
| 8 | MEMOREC | 53994 | 0% | 100% | 2 |
| 9 | UMR 8023 | 53994 | 0% | 100% | 2 |
| 10 | GSF FOR UNGSZENTRUM UMWELT GESUNDHEIT | 40075 | 1% | 21% | 7 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | LECTURE NOTES IN BIOINFORMATICS | 124919 | 2% | 24% | 19 |
| 2 | BIOINFORMATICS | 88737 | 17% | 2% | 187 |
| 3 | NUCLEIC ACIDS RESEARCH | 49688 | 24% | 1% | 269 |
| 4 | BMC BIOINFORMATICS | 14665 | 6% | 1% | 64 |
| 5 | COMPARATIVE AND FUNCTIONAL GENOMICS | 11856 | 1% | 3% | 13 |
| 6 | BIOTECHFORUM ( BTF ) | 6485 | 2% | 1% | 25 |
| 7 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 5794 | 1% | 2% | 14 |
| 8 | INTERNATIONAL JOURNAL OF DATA MINING AND BIOINFORMATICS | 3993 | 1% | 2% | 8 |
| 9 | TRENDS IN GENETICS | 2517 | 1% | 1% | 16 |
| 10 | GENE-COMBIS | 2248 | 0% | 8% | 1 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SIMILARITY SCORE DISSECTION | 80991 | 0% | 100% | 3 | Search SIMILARITY+SCORE+DISSECTION | Search SIMILARITY+SCORE+DISSECTION |
| 2 | INTERPRO | 61704 | 0% | 57% | 4 | Search INTERPRO | Search INTERPRO |
| 3 | PROTEIN SEQUENCE CLUSTERING | 60742 | 0% | 75% | 3 | Search PROTEIN+SEQUENCE+CLUSTERING | Search PROTEIN+SEQUENCE+CLUSTERING |
| 4 | 4D BIOINFORMATICS | 53994 | 0% | 100% | 2 | Search 4D+BIOINFORMATICS | Search 4D+BIOINFORMATICS |
| 5 | ADRIS | 53994 | 0% | 100% | 2 | Search ADRIS | Search ADRIS |
| 6 | BLAST SOFTWARE | 53994 | 0% | 100% | 2 | Search BLAST+SOFTWARE | Search BLAST+SOFTWARE |
| 7 | FOLD CRITICAL SEQUENCE SEGMENT | 53994 | 0% | 100% | 2 | Search FOLD+CRITICAL+SEQUENCE+SEGMENT | Search FOLD+CRITICAL+SEQUENCE+SEGMENT |
| 8 | FOSSIL SEDIMENT FILTRATION | 53994 | 0% | 100% | 2 | Search FOSSIL+SEDIMENT+FILTRATION | Search FOSSIL+SEDIMENT+FILTRATION |
| 9 | MOLECULAR BIOLOGY DATABASE | 53994 | 0% | 100% | 2 | Search MOLECULAR+BIOLOGY+DATABASE | Search MOLECULAR+BIOLOGY+DATABASE |
| 10 | NON GLOBULAR SEQUENCE SEGMENT | 53994 | 0% | 100% | 2 | Search NON+GLOBULAR+SEQUENCE+SEGMENT | Search NON+GLOBULAR+SEQUENCE+SEGMENT |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BAIROCH, A , APWEILER, R , WU, CH , BARKER, WC , BOECKMANN, B , FERRO, S , GASTEIGER, E , HUANG, HZ , LOPEZ, R , MAGRANE, M , ET AL (2005) THE UNIVERSAL PROTEIN RESOURCE (UNIPROT).NUCLEIC ACIDS RESEARCH. VOL. 33. ISSUE . P. D154-D159 | 19 | 73% | 674 |
| 2 | BOECKMANN, B , BAIROCH, A , APWEILER, R , BLATTER, MC , ESTREICHER, A , GASTEIGER, E , MARTIN, MJ , MICHOUD, K , O'DONOVAN, C , PHAN, I , ET AL (2003) THE SWISS-PROT PROTEIN KNOWLEDGEBASE AND ITS SUPPLEMENT TREMBL IN 2003.NUCLEIC ACIDS RESEARCH. VOL. 31. ISSUE 1. P. 365-370 | 15 | 65% | 1745 |
| 3 | APWEILER, R , BAIROCH, A , WU, CH , BARKER, WC , BOECKMANN, B , FERRO, S , GASTEIGER, E , HUANG, HZ , LOPEZ, R , MAGRANE, M , ET AL (2004) UNIPROT: THE UNIVERSAL PROTEIN KNOWLEDGEBASE.NUCLEIC ACIDS RESEARCH. VOL. 32. ISSUE . P. D115-D119 | 15 | 75% | 724 |
| 4 | SHOOP, E , SILVERSTEIN, KAT , JOHNSON, JE , RETZEL, EF , (2001) METAFAM: A UNIFIED CLASSIFICATION OF PROTEIN FAMILIES. II. SCHEMA AND QUERY CAPABILITIES.BIOINFORMATICS. VOL. 17. ISSUE 3. P. 262-271 | 24 | 80% | 8 |
| 5 | MULDER, NJ , APWEILER, R , (2002) TOOLS AND RESOURCES FOR IDENTIFYING PROTEIN FAMILIES, DOMAINS AND MOTIFS.GENOME BIOLOGY. VOL. 3. ISSUE 1. P. - | 23 | 79% | 5 |
| 6 | KRAUSE, A , (2006) LARGE SCALE PROTEIN SEQUENCE CLUSTERING - NOT SOLVED BUT SOLVABLE.CURRENT BIOINFORMATICS. VOL. 1. ISSUE 2. P. 247-254 | 26 | 60% | 2 |
| 7 | ZDOBNOV, EM , APWEILER, R , (2001) INTERPROSCAN - AN INTEGRATION PLATFORM FOR THE SIGNATURE-RECOGNITION METHODS IN INTERPRO.BIOINFORMATICS. VOL. 17. ISSUE 9. P. 847-848 | 8 | 89% | 1416 |
| 8 | MURVAI, J , VLAHOVICEK, K , BARTA, E , CATALETTO, B , PONGOR, S , (2000) THE SBASE PROTEIN DOMAIN LIBRARY, RELEASE 7.0: A COLLECTION OF ANNOTATED PROTEIN SEQUENCE SEGMENTS.NUCLEIC ACIDS RESEARCH. VOL. 28. ISSUE 1. P. 260 -262 | 20 | 83% | 11 |
| 9 | CAMON, E , MAGRANE, M , BARRELL, D , BINNS, D , FLEISCHMANN, W , KERSEY, P , MULDER, N , OINN, T , MASLEN, J , COX, A , ET AL (2003) THE GENE ONTOLOGY ANNOTATION (GOA) PROJECT: IMPLEMENTATION OF GO IN SWISS-PROT, TREMBL, AND INTERPRO.GENOME RESEARCH. VOL. 13. ISSUE 4. P. 662-672 | 17 | 65% | 217 |
| 10 | YONA, G , LINIAL, N , LINIAL, M , (2000) PROTOMAP: AUTOMATIC CLASSIFICATION OF PROTEIN SEQUENCES AND HIERARCHY OF PROTEIN FAMILIES.NUCLEIC ACIDS RESEARCH. VOL. 28. ISSUE 1. P. 49-55 | 16 | 84% | 97 |
Classes with closest relation at Level 1 |