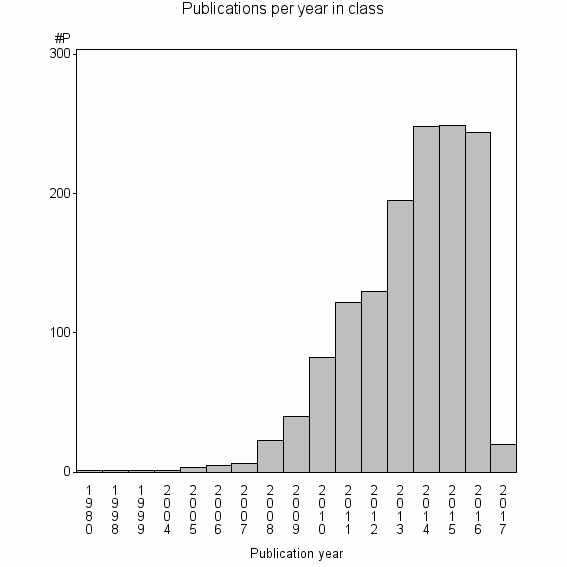
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 7400 | 1371 | 38.7 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 1101 | 2 | BIOINFORMATICS//COPY NUMBER VARIATION//NEXT GENERATION SEQUENCING | 9808 |
| 7400 | 1 | RNA SEQ//TRANSCRIPTOME ASSEMBLY//RNA SEQUENCING | 1371 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA SEQ | authKW | 669080 | 20% | 11% | 268 |
| 2 | TRANSCRIPTOME ASSEMBLY | authKW | 103911 | 1% | 33% | 14 |
| 3 | RNA SEQUENCING | authKW | 82274 | 4% | 8% | 48 |
| 4 | TRANSCRIPTOME | authKW | 82064 | 9% | 3% | 127 |
| 5 | ANIM PROD RD | address | 66812 | 0% | 100% | 3 |
| 6 | DESEQ2 | authKW | 66812 | 0% | 100% | 3 |
| 7 | SPLICING GRAPH | authKW | 66812 | 0% | 100% | 3 |
| 8 | DE NOVO ASSEMBLY | authKW | 64017 | 2% | 11% | 26 |
| 9 | RNA SEQ DATA ANALYSIS | authKW | 59386 | 0% | 67% | 4 |
| 10 | DIFFERENTIAL EXPRESSION ANALYSIS | authKW | 50893 | 1% | 29% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 35631 | 30% | 0% | 406 |
| 2 | Biotechnology & Applied Microbiology | 22194 | 53% | 0% | 733 |
| 3 | Biochemical Research Methods | 12984 | 33% | 0% | 447 |
| 4 | Genetics & Heredity | 10191 | 36% | 0% | 495 |
| 5 | Biochemistry & Molecular Biology | 1078 | 26% | 0% | 361 |
| 6 | Statistics & Probability | 399 | 5% | 0% | 64 |
| 7 | Multidisciplinary Sciences | 306 | 3% | 0% | 38 |
| 8 | Plant Sciences | 36 | 4% | 0% | 54 |
| 9 | Biology | 23 | 2% | 0% | 26 |
| 10 | Evolutionary Biology | 21 | 1% | 0% | 17 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ANIM PROD RD | 66812 | 0% | 100% | 3 |
| 2 | CLIN GENET BIOINFORMAT | 50108 | 0% | 75% | 3 |
| 3 | AGRILIFE GENOM SERV | 44541 | 0% | 100% | 2 |
| 4 | CHAIR BIOINFORMAT GRP | 40085 | 0% | 60% | 3 |
| 5 | GENOM GENE EXP S | 34791 | 0% | 31% | 5 |
| 6 | STATE FO T TREE GENET BREEDING | 29693 | 0% | 67% | 2 |
| 7 | SYST PHARMACOL BIOMARKERS | 29693 | 0% | 67% | 2 |
| 8 | HUB BIOINFORMAT BIOSTAT | 28630 | 0% | 43% | 3 |
| 9 | KOREA ZOONOSIS KOZRI | 28630 | 0% | 43% | 3 |
| 10 | OMICS SCI | 26128 | 1% | 13% | 9 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC GENOMICS | 43647 | 10% | 1% | 137 |
| 2 | BIOINFORMATICS | 43303 | 11% | 1% | 144 |
| 3 | BMC BIOINFORMATICS | 29593 | 7% | 1% | 100 |
| 4 | GENOME BIOLOGY | 16433 | 3% | 2% | 47 |
| 5 | GENOME RESEARCH | 13575 | 2% | 2% | 29 |
| 6 | NATURE METHODS | 8187 | 2% | 1% | 25 |
| 7 | STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY | 6327 | 1% | 2% | 12 |
| 8 | JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY | 4911 | 1% | 2% | 9 |
| 9 | BRIEFINGS IN BIOINFORMATICS | 3856 | 1% | 2% | 11 |
| 10 | NUCLEIC ACIDS RESEARCH | 3596 | 6% | 0% | 81 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA SEQ | 669080 | 20% | 11% | 268 | Search RNA+SEQ | Search RNA+SEQ |
| 2 | TRANSCRIPTOME ASSEMBLY | 103911 | 1% | 33% | 14 | Search TRANSCRIPTOME+ASSEMBLY | Search TRANSCRIPTOME+ASSEMBLY |
| 3 | RNA SEQUENCING | 82274 | 4% | 8% | 48 | Search RNA+SEQUENCING | Search RNA+SEQUENCING |
| 4 | TRANSCRIPTOME | 82064 | 9% | 3% | 127 | Search TRANSCRIPTOME | Search TRANSCRIPTOME |
| 5 | DESEQ2 | 66812 | 0% | 100% | 3 | Search DESEQ2 | Search DESEQ2 |
| 6 | SPLICING GRAPH | 66812 | 0% | 100% | 3 | Search SPLICING+GRAPH | Search SPLICING+GRAPH |
| 7 | DE NOVO ASSEMBLY | 64017 | 2% | 11% | 26 | Search DE+NOVO+ASSEMBLY | Search DE+NOVO+ASSEMBLY |
| 8 | RNA SEQ DATA ANALYSIS | 59386 | 0% | 67% | 4 | Search RNA+SEQ+DATA+ANALYSIS | Search RNA+SEQ+DATA+ANALYSIS |
| 9 | DIFFERENTIAL EXPRESSION ANALYSIS | 50893 | 1% | 29% | 8 | Search DIFFERENTIAL+EXPRESSION+ANALYSIS | Search DIFFERENTIAL+EXPRESSION+ANALYSIS |
| 10 | RNA SEQ DATA | 47154 | 0% | 35% | 6 | Search RNA+SEQ+DATA | Search RNA+SEQ+DATA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | TRAPNELL, C , ROBERTS, A , GOFF, L , PERTEA, G , KIM, D , KELLEY, DR , PIMENTEL, H , SALZBERG, SL , RINN, JL , PACHTER, L , (2012) DIFFERENTIAL GENE AND TRANSCRIPT EXPRESSION ANALYSIS OF RNA-SEQ EXPERIMENTS WITH TOPHAT AND CUFFLINKS.NATURE PROTOCOLS. VOL. 7. ISSUE 3. P. 562 -578 | 29 | 71% | 2089 |
| 2 | CONESA, A , MADRIGAL, P , TARAZONA, S , GOMEZ-CABRERO, D , CERVERA, A , MCPHERSON, A , SZCZESNIAK, MW , GAFFNEY, DJ , ELO, LL , ZHANG, XG , ET AL (2016) A SURVEY OF BEST PRACTICES FOR RNA-SEQ DATA ANALYSIS.GENOME BIOLOGY. VOL. 17. ISSUE . P. - | 86 | 45% | 23 |
| 3 | LI, B , DEWEY, CN , (2011) RSEM: ACCURATE TRANSCRIPT QUANTIFICATION FROM RNA-SEQ DATA WITH OR WITHOUT A REFERENCE GENOME.BMC BIOINFORMATICS. VOL. 12. ISSUE . P. - | 29 | 76% | 1329 |
| 4 | HAAS, BJ , PAPANICOLAOU, A , YASSOUR, M , GRABHERR, M , BLOOD, PD , BOWDEN, J , COUGER, MB , ECCLES, D , LI, B , LIEBER, M , ET AL (2013) DE NOVO TRANSCRIPT SEQUENCE RECONSTRUCTION FROM RNA-SEQ USING THE TRINITY PLATFORM FOR REFERENCE GENERATION AND ANALYSIS.NATURE PROTOCOLS. VOL. 8. ISSUE 8. P. 1494 -1512 | 29 | 67% | 734 |
| 5 | SONESON, C , DELORENZI, M , (2013) A COMPARISON OF METHODS FOR DIFFERENTIAL EXPRESSION ANALYSIS OF RNA-SEQ DATA.BMC BIOINFORMATICS. VOL. 14. ISSUE . P. - | 26 | 96% | 182 |
| 6 | ZHENG, CL , KAWANE, S , BOTTOMLY, D , WILMOT, B , (2014) ANALYSIS CONSIDERATIONS FOR UTILIZING RNA-SEQ TO CHARACTERIZE THE BRAIN TRANSCRIPTOME.BRAIN TRANSCRIPTOME. VOL. 116. ISSUE . P. 21 -54 | 80 | 50% | 0 |
| 7 | FINOTELLO, F , DI CAMILLO, B , (2015) MEASURING DIFFERENTIAL GENE EXPRESSION WITH RNA-SEQ: CHALLENGES AND STRATEGIES FOR DATA ANALYSIS.BRIEFINGS IN FUNCTIONAL GENOMICS. VOL. 14. ISSUE 2. P. 130 -142 | 49 | 68% | 10 |
| 8 | ANDERS, S , MCCARTHY, DJ , CHEN, YS , OKONIEWSKI, M , SMYTH, GK , HUBER, W , ROBINSON, MD , (2013) COUNT-BASED DIFFERENTIAL EXPRESSION ANALYSIS OF RNA SEQUENCING DATA USING R AND BIOCONDUCTOR.NATURE PROTOCOLS. VOL. 8. ISSUE 9. P. 1765-1786 | 34 | 63% | 203 |
| 9 | LOVE, MI , HUBER, W , ANDERS, S , (2014) MODERATED ESTIMATION OF FOLD CHANGE AND DISPERSION FOR RNA-SEQ DATA WITH DESEQ2.GENOME BIOLOGY. VOL. 15. ISSUE 12. P. - | 24 | 60% | 297 |
| 10 | TANG, M , SUN, JQ , SHIMIZU, K , KADOTA, K , (2015) EVALUATION OF METHODS FOR DIFFERENTIAL EXPRESSION ANALYSIS ON MULTI-GROUP RNA-SEQ COUNT DATA.BMC BIOINFORMATICS. VOL. 16. ISSUE . P. - | 41 | 87% | 1 |
Classes with closest relation at Level 1 |