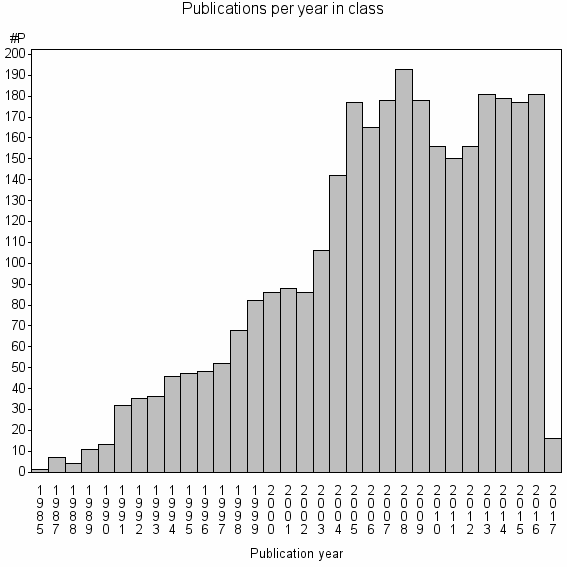
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 785 | 3077 | 52.7 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 785 | 1 | SR PROTEINS//ALTERNATIVE SPLICING//POLYPYRIMIDINE TRACT BINDING PROTEIN | 3077 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SR PROTEINS | authKW | 890577 | 5% | 61% | 148 |
| 2 | ALTERNATIVE SPLICING | authKW | 634338 | 18% | 12% | 548 |
| 3 | POLYPYRIMIDINE TRACT BINDING PROTEIN | authKW | 200887 | 1% | 56% | 36 |
| 4 | SPLICING FACTOR | authKW | 180388 | 1% | 43% | 42 |
| 5 | SRPK1 | authKW | 174945 | 1% | 77% | 23 |
| 6 | RS DOMAIN | authKW | 164008 | 1% | 72% | 23 |
| 7 | SPLICING ENHANCER | authKW | 145780 | 1% | 64% | 23 |
| 8 | PRE MRNA SPLICING | authKW | 143338 | 3% | 18% | 82 |
| 9 | SPLICING REGULATION | authKW | 101736 | 1% | 49% | 21 |
| 10 | U2AF | authKW | 99477 | 1% | 53% | 19 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 15782 | 59% | 0% | 1806 |
| 2 | Cell Biology | 10961 | 31% | 0% | 962 |
| 3 | Genetics & Heredity | 9021 | 23% | 0% | 716 |
| 4 | Biotechnology & Applied Microbiology | 1603 | 11% | 0% | 337 |
| 5 | Mathematical & Computational Biology | 1486 | 4% | 0% | 133 |
| 6 | Developmental Biology | 1046 | 4% | 0% | 125 |
| 7 | Biochemical Research Methods | 561 | 5% | 0% | 167 |
| 8 | Biophysics | 357 | 5% | 0% | 154 |
| 9 | Oncology | 113 | 5% | 0% | 166 |
| 10 | Evolutionary Biology | 55 | 1% | 0% | 40 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA RNP GRP | 79375 | 0% | 100% | 8 |
| 2 | PROGRAM MOL PLANT BIOL | 61730 | 1% | 33% | 19 |
| 3 | GENOM PHYSIOL | 50222 | 0% | 56% | 9 |
| 4 | EQUIPE BASES MOL PROG S CANC POUMON 2 | 35432 | 0% | 71% | 5 |
| 5 | PROGRAM MOL CANC BIOL | 30378 | 0% | 44% | 7 |
| 6 | MICROBIOL INFECTIOL | 26446 | 1% | 10% | 27 |
| 7 | CORE MOL PHARMACOL | 22323 | 0% | 75% | 3 |
| 8 | HAVANA GRP | 22323 | 0% | 75% | 3 |
| 9 | HUGHES UNDER | 22323 | 0% | 75% | 3 |
| 10 | EXONHIT | 19844 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 29504 | 3% | 3% | 87 |
| 2 | RNA-A PUBLICATION OF THE RNA SOCIETY | 18649 | 2% | 4% | 49 |
| 3 | NUCLEIC ACIDS RESEARCH | 17257 | 9% | 1% | 264 |
| 4 | MOLECULAR AND CELLULAR BIOLOGY | 13935 | 6% | 1% | 178 |
| 5 | GENES & DEVELOPMENT | 10678 | 3% | 1% | 90 |
| 6 | GENOME BIOLOGY | 6661 | 1% | 2% | 45 |
| 7 | MOLECULAR CELL | 6239 | 2% | 1% | 59 |
| 8 | BIOTECHFORUM ( BTF ) | 4639 | 1% | 1% | 35 |
| 9 | RNA BIOLOGY | 4289 | 1% | 2% | 20 |
| 10 | BMC GENOMICS | 3885 | 2% | 1% | 62 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SR PROTEINS | 890577 | 5% | 61% | 148 | Search SR+PROTEINS | Search SR+PROTEINS |
| 2 | ALTERNATIVE SPLICING | 634338 | 18% | 12% | 548 | Search ALTERNATIVE+SPLICING | Search ALTERNATIVE+SPLICING |
| 3 | POLYPYRIMIDINE TRACT BINDING PROTEIN | 200887 | 1% | 56% | 36 | Search POLYPYRIMIDINE+TRACT+BINDING+PROTEIN | Search POLYPYRIMIDINE+TRACT+BINDING+PROTEIN |
| 4 | SPLICING FACTOR | 180388 | 1% | 43% | 42 | Search SPLICING+FACTOR | Search SPLICING+FACTOR |
| 5 | SRPK1 | 174945 | 1% | 77% | 23 | Search SRPK1 | Search SRPK1 |
| 6 | RS DOMAIN | 164008 | 1% | 72% | 23 | Search RS+DOMAIN | Search RS+DOMAIN |
| 7 | SPLICING ENHANCER | 145780 | 1% | 64% | 23 | Search SPLICING+ENHANCER | Search SPLICING+ENHANCER |
| 8 | PRE MRNA SPLICING | 143338 | 3% | 18% | 82 | Search PRE+MRNA+SPLICING | Search PRE+MRNA+SPLICING |
| 9 | SPLICING REGULATION | 101736 | 1% | 49% | 21 | Search SPLICING+REGULATION | Search SPLICING+REGULATION |
| 10 | U2AF | 99477 | 1% | 53% | 19 | Search U2AF | Search U2AF |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BLACK, DL , (2003) MECHANISMS OF ALTERNATIVE PRE-MESSENGER RNA SPLICING.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 72. ISSUE . P. 291-336 | 184 | 52% | 1375 |
| 2 | LONG, JC , CACERES, JF , (2009) THE SR PROTEIN FAMILY OF SPLICING FACTORS: MASTER REGULATORS OF GENE EXPRESSION.BIOCHEMICAL JOURNAL. VOL. 417. ISSUE . P. 15-27 | 138 | 67% | 440 |
| 3 | ZHOU, ZH , FU, XD , (2013) REGULATION OF SPLICING BY SR PROTEINS AND SR PROTEIN-SPECIFIC KINASES.CHROMOSOMA. VOL. 122. ISSUE 3. P. 191 -207 | 144 | 75% | 67 |
| 4 | FU, XD , ARES, M , (2014) CONTEXT-DEPENDENT CONTROL OF ALTERNATIVE SPLICING BY RNA-BINDING PROTEINS.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 10. P. 689 -701 | 121 | 71% | 90 |
| 5 | MATLIN, AJ , CLARK, F , SMITH, CWJ , (2005) UNDERSTANDING ALTERNATIVE SPLICING: TOWARDS A CELLULAR CODE.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 6. ISSUE 5. P. 386-398 | 113 | 84% | 662 |
| 6 | LIN, SR , FU, XD , (2007) SR PROTEINS AND RELATED FACTORS IN ALTERNATIVE SPLICING.ALTERNATIVE SPLICING IN THE POSTGENOMIC ERA. VOL. 623. ISSUE . P. 107-122 | 140 | 85% | 102 |
| 7 | GRAVELEY, BR , (2000) SORTING OUT THE COMPLEXITY OF SR PROTEIN FUNCTIONS.RNA. VOL. 6. ISSUE 9. P. 1197-1211 | 113 | 82% | 734 |
| 8 | HOWARD, JM , SANFORD, JR , (2015) THE RNAISSANCE FAMILY: SR PROTEINS AS MULTIFACETED REGULATORS OF GENE EXPRESSION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 6. ISSUE 1. P. 93 -110 | 129 | 71% | 12 |
| 9 | CHEN, M , MANLEY, JL , (2009) MECHANISMS OF ALTERNATIVE SPLICING REGULATION: INSIGHTS FROM MOLECULAR AND GENOMICS APPROACHES.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 10. ISSUE 11. P. 741 -754 | 103 | 64% | 437 |
| 10 | KEPPETIPOLA, N , SHARMA, S , LI, Q , BLACK, DL , (2012) NEURONAL REGULATION OF PRE-MRNA SPLICING BY POLYPYRIMIDINE TRACT BINDING PROTEINS, PTBP1 AND PTBP2.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 47. ISSUE 4. P. 360-378 | 122 | 77% | 31 |
Classes with closest relation at Level 1 |