Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
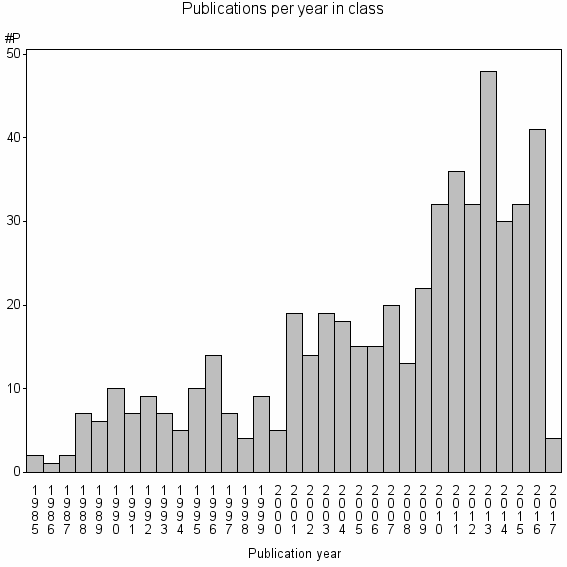
| 19243 | 515 | 38.8 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 1101 | 2 | BIOINFORMATICS//COPY NUMBER VARIATION//NEXT GENERATION SEQUENCING | 9808 |
| 19243 | 1 | TRANS SPLICING//SPLICED LEADER//CHIMERIC RNA | 515 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | TRANS SPLICING | authKW | 1067341 | 14% | 25% | 71 |
| 2 | SPLICED LEADER | authKW | 809481 | 6% | 43% | 32 |
| 3 | CHIMERIC RNA | authKW | 282327 | 2% | 48% | 10 |
| 4 | RNA TRANS SPLICING | authKW | 135519 | 1% | 57% | 4 |
| 5 | PROGRAM MOL THER Y GENODERMATOSES | address | 133403 | 1% | 75% | 3 |
| 6 | SPLICE LEADER | authKW | 133403 | 1% | 75% | 3 |
| 7 | BIOLOSITICS | authKW | 118582 | 0% | 100% | 2 |
| 8 | CANCER SPECIFIC FUSION GENES | authKW | 118582 | 0% | 100% | 2 |
| 9 | CONJOINED GENES | authKW | 118582 | 0% | 100% | 2 |
| 10 | HETEROLOGOUS TRANS SPLICING | authKW | 118582 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 2941 | 32% | 0% | 164 |
| 2 | Biochemistry & Molecular Biology | 1889 | 50% | 0% | 260 |
| 3 | Biotechnology & Applied Microbiology | 1303 | 22% | 0% | 114 |
| 4 | Parasitiology | 750 | 8% | 0% | 39 |
| 5 | Cell Biology | 654 | 20% | 0% | 101 |
| 6 | Mathematical & Computational Biology | 653 | 7% | 0% | 35 |
| 7 | Oncology | 248 | 13% | 0% | 68 |
| 8 | Biochemical Research Methods | 208 | 8% | 0% | 39 |
| 9 | Medicine, Research & Experimental | 95 | 6% | 0% | 32 |
| 10 | Developmental Biology | 31 | 2% | 0% | 10 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROGRAM MOL THER Y GENODERMATOSES | 133403 | 1% | 75% | 3 |
| 2 | MOL DERMATOL EB HOUSE AUSTRIA | 106721 | 1% | 60% | 3 |
| 3 | AGR INFORMAT SUB | 59291 | 0% | 100% | 1 |
| 4 | AGR LIGE SCI | 59291 | 0% | 100% | 1 |
| 5 | ANIM SCI S | 59291 | 0% | 100% | 1 |
| 6 | ATLAS POITIERS EDITORIAL GRP | 59291 | 0% | 100% | 1 |
| 7 | BIOCHEM BIOPHYS SCI 3201 CULLEN BLVD | 59291 | 0% | 100% | 1 |
| 8 | BIOL DEV UMR 7138 | 59291 | 0% | 100% | 1 |
| 9 | BIOL MC 4616 | 59291 | 0% | 100% | 1 |
| 10 | CIHR MSFHR BIOINFORMAT TRAINING PROGRAM | 59291 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND BIOCHEMICAL PARASITOLOGY | 6496 | 5% | 0% | 25 |
| 2 | RNA-A PUBLICATION OF THE RNA SOCIETY | 5623 | 2% | 1% | 11 |
| 3 | GENOME RESEARCH | 3477 | 2% | 1% | 9 |
| 4 | BIOTECHFORUM ( BTF ) | 3283 | 2% | 0% | 12 |
| 5 | NUCLEIC ACIDS RESEARCH | 2481 | 8% | 0% | 41 |
| 6 | BIOINFORMATICS | 2199 | 4% | 0% | 20 |
| 7 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 1448 | 1% | 1% | 3 |
| 8 | BMC GENOMICS | 1193 | 3% | 0% | 14 |
| 9 | RNA | 1134 | 1% | 0% | 7 |
| 10 | MOLECULAR THERAPY | 1057 | 2% | 0% | 8 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRANS SPLICING | 1067341 | 14% | 25% | 71 | Search TRANS+SPLICING | Search TRANS+SPLICING |
| 2 | SPLICED LEADER | 809481 | 6% | 43% | 32 | Search SPLICED+LEADER | Search SPLICED+LEADER |
| 3 | CHIMERIC RNA | 282327 | 2% | 48% | 10 | Search CHIMERIC+RNA | Search CHIMERIC+RNA |
| 4 | RNA TRANS SPLICING | 135519 | 1% | 57% | 4 | Search RNA+TRANS+SPLICING | Search RNA+TRANS+SPLICING |
| 5 | SPLICE LEADER | 133403 | 1% | 75% | 3 | Search SPLICE+LEADER | Search SPLICE+LEADER |
| 6 | BIOLOSITICS | 118582 | 0% | 100% | 2 | Search BIOLOSITICS | Search BIOLOSITICS |
| 7 | CANCER SPECIFIC FUSION GENES | 118582 | 0% | 100% | 2 | Search CANCER+SPECIFIC+FUSION+GENES | Search CANCER+SPECIFIC+FUSION+GENES |
| 8 | CONJOINED GENES | 118582 | 0% | 100% | 2 | Search CONJOINED+GENES | Search CONJOINED+GENES |
| 9 | HETEROLOGOUS TRANS SPLICING | 118582 | 0% | 100% | 2 | Search HETEROLOGOUS+TRANS+SPLICING | Search HETEROLOGOUS+TRANS+SPLICING |
| 10 | SPLICED LEADER SEQUENCE RNA | 118582 | 0% | 100% | 2 | Search SPLICED+LEADER+SEQUENCE+RNA | Search SPLICED+LEADER+SEQUENCE+RNA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LASDA, EL , BLUMENTHAL, T , (2011) TRANS-SPLICING.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 2. ISSUE 3. P. 417-434 | 79 | 58% | 36 |
| 2 | LATYSHEVA, NS , BABU, MM , (2016) DISCOVERING AND UNDERSTANDING ONCOGENIC GENE FUSIONS THROUGH DATA INTENSIVE COMPUTATIONAL APPROACHES.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 10. P. 4487 -4503 | 81 | 47% | 2 |
| 3 | LEI, Q , LI, C , ZUO, ZX , HUANG, CH , CHENG, HH , ZHOU, RJ , (2016) EVOLUTIONARY INSIGHTS INTO RNA TRANS-SPLICING IN VERTEBRATES.GENOME BIOLOGY AND EVOLUTION. VOL. 8. ISSUE 3. P. 562 -577 | 68 | 52% | 0 |
| 4 | KUMAR, S , RAZZAQ, SK , VO, AD , GAUTAM, M , LI, H , (2016) IDENTIFYING FUSION TRANSCRIPTS USING NEXT GENERATION SEQUENCING.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 7. ISSUE 6. P. 811 -823 | 47 | 67% | 0 |
| 5 | RODRIGUEZ-MARTIN, B , PALUMBO, E , MARCO-SOLA, S , GRIEBEL, T , RIBECA, P , ALONSO, G , RASTROJO, A , AGUADO, B , GUIGO, R , DJEBALI, S , (2017) CHIMPIPE: ACCURATE DETECTION OF FUSION GENES AND TRANSCRIPTION-INDUCED CHIMERAS FROM RNA-SEQ DATA.BMC GENOMICS. VOL. 18. ISSUE . P. - | 38 | 69% | 0 |
| 6 | YU, CY , LIU, HJ , HUNG, LY , KUO, HC , CHUANG, TJ , (2014) IS AN OBSERVED NON-CO-LINEAR RNA PRODUCT SPLICED IN TRANS, IN CIS OR JUST IN VITRO?.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE 14. P. 9410 -9423 | 37 | 67% | 9 |
| 7 | PETTITT, J , HARRISON, N , STANSFIELD, I , CONNOLLY, B , MULLER, B , (2010) THE EVOLUTION OF SPLICED LEADER TRANS-SPLICING IN NEMATODES.BIOCHEMICAL SOCIETY TRANSACTIONS. VOL. 38. ISSUE . P. 1125-1130 | 35 | 65% | 7 |
| 8 | CHUANG, TJ , WU, CS , CHEN, CY , HUNG, LY , CHIANG, TW , YANG, MY , (2016) NCLSCAN: ACCURATE IDENTIFICATION OF NON-CO-LINEAR TRANSCRIPTS (FUSION, TRANS-SPLICING AND CIRCULAR RNA) WITH A GOOD BALANCE BETWEEN SENSITIVITY AND PRECISION.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 3. P. - | 42 | 44% | 1 |
| 9 | MORTON, JJ , BLUMENTHAL, T , (2011) RNA PROCESSING IN C. ELEGANS.CAENORHABDITIS ELEGANS: MOLECULAR GENETICS AND DEVELOPMENT, SECOND EDITION. VOL. 106. ISSUE . P. 187 -217 | 54 | 40% | 6 |
| 10 | LIU, S , TSAI, WH , DING, Y , CHEN, R , FANG, Z , HUO, ZG , KIM, S , MA, TZ , CHANG, TY , PRIEDIGKEIT, NM , ET AL (2016) COMPREHENSIVE EVALUATION OF FUSION TRANSCRIPT DETECTION ALGORITHMS AND A META-CALLER TO COMBINE TOP PERFORMING METHODS IN PAIRED-END RNA-SEQ DATA.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 5. P. - | 26 | 51% | 6 |
Classes with closest relation at Level 1 |