Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
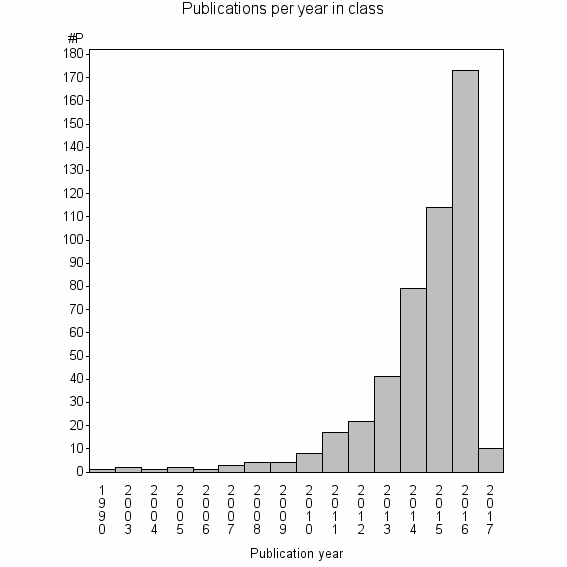
| 19974 | 482 | 46.0 | 91% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 1101 | 2 | BIOINFORMATICS//COPY NUMBER VARIATION//NEXT GENERATION SEQUENCING | 9808 |
| 19974 | 1 | SINGLE CELL RNA SEQ//SINGLE CELL RNA SEQUENCING//SINGLE CELL TRANSCRIPTOMICS | 482 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SINGLE CELL RNA SEQ | authKW | 348416 | 2% | 50% | 11 |
| 2 | SINGLE CELL RNA SEQUENCING | authKW | 285074 | 1% | 75% | 6 |
| 3 | SINGLE CELL TRANSCRIPTOMICS | authKW | 228056 | 1% | 60% | 6 |
| 4 | SINGLE CELL BIOLOGY | authKW | 152034 | 1% | 40% | 6 |
| 5 | SINGLE CELL GENE EXPRESSION PROFILING | authKW | 142537 | 1% | 75% | 3 |
| 6 | SANGER EBI | address | 126701 | 0% | 100% | 2 |
| 7 | UEA FLOW CYTOMETRY SERV | address | 126701 | 0% | 100% | 2 |
| 8 | SINGLE CELL | authKW | 113219 | 7% | 5% | 33 |
| 9 | SINGLE CELL GENOM | address | 105577 | 1% | 33% | 5 |
| 10 | SINGLE CELL TRANSCRIPTOME | authKW | 95023 | 1% | 50% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemical Research Methods | 4738 | 33% | 0% | 160 |
| 2 | Biotechnology & Applied Microbiology | 3007 | 34% | 0% | 163 |
| 3 | Mathematical & Computational Biology | 1413 | 10% | 0% | 49 |
| 4 | Genetics & Heredity | 1355 | 23% | 0% | 110 |
| 5 | Cell Biology | 1063 | 25% | 0% | 121 |
| 6 | Biochemistry & Molecular Biology | 528 | 30% | 0% | 145 |
| 7 | Multidisciplinary Sciences | 139 | 3% | 0% | 15 |
| 8 | Cell & Tissue Engineering | 105 | 1% | 0% | 7 |
| 9 | Developmental Biology | 65 | 3% | 0% | 13 |
| 10 | Chemistry, Analytical | 27 | 5% | 0% | 23 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SANGER EBI | 126701 | 0% | 100% | 2 |
| 2 | UEA FLOW CYTOMETRY SERV | 126701 | 0% | 100% | 2 |
| 3 | SINGLE CELL GENOM | 105577 | 1% | 33% | 5 |
| 4 | BEIJING ADV INNOVAT GENOM ICG | 84466 | 0% | 67% | 2 |
| 5 | KLARMAN CELL OBSERV | 84466 | 0% | 67% | 2 |
| 6 | EBI SINGLE CELL GENOM | 81447 | 1% | 43% | 3 |
| 7 | BIO ABORAT BIO 5 | 63350 | 0% | 100% | 1 |
| 8 | BIOL COMP INFORMAT SCI | 63350 | 0% | 100% | 1 |
| 9 | BIOSYSTE SCI ENGN | 63350 | 0% | 100% | 1 |
| 10 | CANC SIGNALING 1 | 63350 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NATURE METHODS | 31468 | 6% | 2% | 29 |
| 2 | GENOME BIOLOGY | 15460 | 6% | 1% | 27 |
| 3 | NATURE BIOTECHNOLOGY | 7813 | 4% | 1% | 18 |
| 4 | CELL SYSTEMS | 5584 | 1% | 3% | 3 |
| 5 | NATURE PROTOCOLS | 4014 | 2% | 1% | 12 |
| 6 | BIOINFORMATICS | 2850 | 5% | 0% | 22 |
| 7 | GENOME RESEARCH | 2245 | 1% | 1% | 7 |
| 8 | GIGASCIENCE | 1732 | 0% | 1% | 2 |
| 9 | PLOS COMPUTATIONAL BIOLOGY | 1354 | 2% | 0% | 10 |
| 10 | BMC MOLECULAR BIOLOGY | 1331 | 1% | 1% | 4 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BACHER, R , KENDZIORSKI, C , (2016) DESIGN AND COMPUTATIONAL ANALYSIS OF SINGLE-CELL RNA-SEQUENCING EXPERIMENTS.GENOME BIOLOGY. VOL. 17. ISSUE . P. - | 58 | 69% | 6 |
| 2 | WEN, L , TANG, FC , (2016) SINGLE-CELL SEQUENCING IN STEM CELL BIOLOGY.GENOME BIOLOGY. VOL. 17. ISSUE . P. - | 61 | 60% | 6 |
| 3 | KUMAR, P , TAN, YQ , CAHAN, P , (2017) UNDERSTANDING DEVELOPMENT AND STEM CELLS USING SINGLE CELL-BASED ANALYSES OF GENE EXPRESSION.DEVELOPMENT. VOL. 144. ISSUE 1. P. 17 -32 | 74 | 58% | 0 |
| 4 | WAGNER, A , REGEV, A , YOSEF, N , (2016) REVEALING THE VECTORS OF CELLULAR IDENTITY WITH SINGLE-CELL GENOMICS.NATURE BIOTECHNOLOGY. VOL. 34. ISSUE 11. P. 1145 -1160 | 91 | 44% | 0 |
| 5 | KOLODZIEJCZYK, AA , KIM, JK , SVENSSON, V , MARIONI, JC , TEICHMANN, SA , (2015) THE TECHNOLOGY AND BIOLOGY OF SINGLE-CELL RNA SEQUENCING.MOLECULAR CELL. VOL. 58. ISSUE 4. P. 610 -620 | 45 | 55% | 42 |
| 6 | TRAPNELL, C , (2015) DEFINING CELL TYPES AND STATES WITH SINGLE-CELL GENOMICS.GENOME RESEARCH. VOL. 25. ISSUE 10. P. 1491 -1498 | 35 | 52% | 39 |
| 7 | POULIN, JF , TASIC, B , HJERLING-LEFFLER, J , TRIMARCHI, JM , AWATRAMANI, R , (2016) DISENTANGLING NEURAL CELL DIVERSITY USING SINGLE-CELL TRANSCRIPTOMICS.NATURE NEUROSCIENCE. VOL. 19. ISSUE 9. P. 1131 -1141 | 55 | 40% | 3 |
| 8 | SAADATPOUR, A , LAI, SJ , GUO, GJ , YUAN, GC , (2015) SINGLE-CELL ANALYSIS IN CANCER GENOMICS.TRENDS IN GENETICS. VOL. 31. ISSUE 10. P. 576 -586 | 45 | 49% | 9 |
| 9 | GRUN, D , VAN OUDENAARDEN, A , (2015) DESIGN AND ANALYSIS OF SINGLE-CELL SEQUENCING EXPERIMENTS.CELL. VOL. 163. ISSUE 4. P. 799 -810 | 35 | 49% | 26 |
| 10 | ZURAUSKIENE, J , YAU, C , (2016) PCAREDUCE: HIERARCHICAL CLUSTERING OF SINGLE CELL TRANSCRIPTIONAL PROFILES.BMC BIOINFORMATICS. VOL. 17. ISSUE . P. - | 23 | 88% | 1 |
Classes with closest relation at Level 1 |