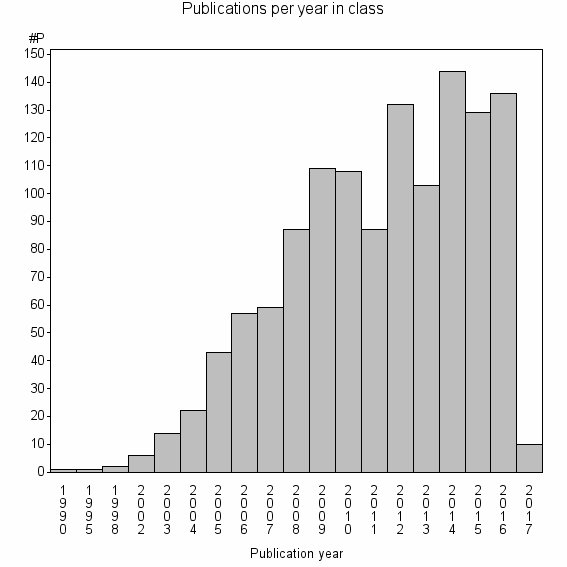
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8478 | 1250 | 46.6 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 447 | 3 | TCTP//TRANSLATIONALLY CONTROLLED TUMOR PROTEIN//GENETIC EPIDEMIOLOGY | 26092 |
| 257 | 2 | GENETIC EPIDEMIOLOGY//HUMAN HEREDITY//GENETICS & HEREDITY | 19642 |
| 8478 | 1 | EQTL//GENETICAL GENOMICS//EQTL MAPPING | 1250 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | EQTL | authKW | 420813 | 4% | 31% | 56 |
| 2 | GENETICAL GENOMICS | authKW | 244927 | 2% | 53% | 19 |
| 3 | EQTL MAPPING | authKW | 203552 | 1% | 83% | 10 |
| 4 | EXPRESSION QUANTITATIVE TRAIT LOCI | authKW | 176463 | 1% | 43% | 17 |
| 5 | GRONINGEN BIOINFORMAT | address | 103484 | 2% | 16% | 27 |
| 6 | GENET MED DEV | address | 90152 | 4% | 7% | 51 |
| 7 | ALLELE SPECIFIC EXPRESSION | authKW | 76704 | 1% | 18% | 17 |
| 8 | EXPRESSION QUANTITATIVE TRAIT LOCUS | authKW | 71049 | 1% | 36% | 8 |
| 9 | LYMPHOBLASTOID CELL LINES | authKW | 70235 | 1% | 18% | 16 |
| 10 | EXPRESSION QUANTITATIVE TRAIT LOCI EQTL | authKW | 67847 | 0% | 56% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 26546 | 60% | 0% | 749 |
| 2 | Mathematical & Computational Biology | 5302 | 12% | 0% | 152 |
| 3 | Biotechnology & Applied Microbiology | 3551 | 23% | 0% | 292 |
| 4 | Biochemical Research Methods | 1130 | 11% | 0% | 135 |
| 5 | Biochemistry & Molecular Biology | 420 | 19% | 0% | 239 |
| 6 | Statistics & Probability | 72 | 2% | 0% | 30 |
| 7 | Biology | 44 | 2% | 0% | 30 |
| 8 | Plant Sciences | 40 | 4% | 0% | 52 |
| 9 | Evolutionary Biology | 11 | 1% | 0% | 13 |
| 10 | Horticulture | 10 | 1% | 0% | 9 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GRONINGEN BIOINFORMAT | 103484 | 2% | 16% | 27 |
| 2 | GENET MED DEV | 90152 | 4% | 7% | 51 |
| 3 | ESTONIAN GENOME PROJECT FDN | 48853 | 0% | 100% | 2 |
| 4 | GBIC | 48853 | 0% | 100% | 2 |
| 5 | COMM CLIN PHARMACOL PHARMACOGENOM | 37972 | 1% | 11% | 14 |
| 6 | GENET GENOM GENEVA | 34727 | 1% | 18% | 8 |
| 7 | GENOM COORDINAT | 33520 | 1% | 15% | 9 |
| 8 | ICAHN GENOM MULTISCALE BIOL | 32883 | 1% | 9% | 15 |
| 9 | FUNCT GENOM GENET PROGRAM | 32568 | 0% | 67% | 2 |
| 10 | MOL NEUROSCI NEURODEGENERAT DIS | 32568 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PLOS GENETICS | 35040 | 7% | 2% | 93 |
| 2 | GENOME RESEARCH | 8561 | 2% | 2% | 22 |
| 3 | GENOME MEDICINE | 8374 | 1% | 2% | 15 |
| 4 | BIOINFORMATICS | 7378 | 5% | 1% | 57 |
| 5 | BMC GENOMICS | 6574 | 4% | 1% | 51 |
| 6 | NATURE GENETICS | 6350 | 3% | 1% | 36 |
| 7 | PHARMACOGENOMICS | 5484 | 2% | 1% | 20 |
| 8 | GENOME BIOLOGY | 5079 | 2% | 1% | 25 |
| 9 | HUMAN MOLECULAR GENETICS | 5014 | 4% | 0% | 45 |
| 10 | BIOTECHFORUM ( BTF ) | 4535 | 2% | 1% | 22 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EQTL | 420813 | 4% | 31% | 56 | Search EQTL | Search EQTL |
| 2 | GENETICAL GENOMICS | 244927 | 2% | 53% | 19 | Search GENETICAL+GENOMICS | Search GENETICAL+GENOMICS |
| 3 | EQTL MAPPING | 203552 | 1% | 83% | 10 | Search EQTL+MAPPING | Search EQTL+MAPPING |
| 4 | EXPRESSION QUANTITATIVE TRAIT LOCI | 176463 | 1% | 43% | 17 | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCI | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCI |
| 5 | ALLELE SPECIFIC EXPRESSION | 76704 | 1% | 18% | 17 | Search ALLELE+SPECIFIC+EXPRESSION | Search ALLELE+SPECIFIC+EXPRESSION |
| 6 | EXPRESSION QUANTITATIVE TRAIT LOCUS | 71049 | 1% | 36% | 8 | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCUS | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCUS |
| 7 | LYMPHOBLASTOID CELL LINES | 70235 | 1% | 18% | 16 | Search LYMPHOBLASTOID+CELL+LINES | Search LYMPHOBLASTOID+CELL+LINES |
| 8 | EXPRESSION QUANTITATIVE TRAIT LOCI EQTL | 67847 | 0% | 56% | 5 | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCI+EQTL | Search EXPRESSION+QUANTITATIVE+TRAIT+LOCI+EQTL |
| 9 | ESNPS | 48853 | 0% | 100% | 2 | Search ESNPS | Search ESNPS |
| 10 | EXPRESSION PHENOTYPE | 48853 | 0% | 100% | 2 | Search EXPRESSION+PHENOTYPE | Search EXPRESSION+PHENOTYPE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ALBERT, FW , KRUGLYAK, L , (2015) THE ROLE OF REGULATORY VARIATION IN COMPLEX TRAITS AND DISEASE.NATURE REVIEWS GENETICS. VOL. 16. ISSUE 4. P. 197 -212 | 127 | 58% | 76 |
| 2 | GAFFNEY, DJ , (2013) GLOBAL PROPERTIES AND FUNCTIONAL COMPLEXITY OF HUMAN GENE REGULATORY VARIATION.PLOS GENETICS. VOL. 9. ISSUE 5. P. - | 64 | 69% | 22 |
| 3 | STRANGER, BE , RAJ, T , (2013) GENETICS OF HUMAN GENE EXPRESSION.CURRENT OPINION IN GENETICS & DEVELOPMENT. VOL. 23. ISSUE 6. P. 627-634 | 52 | 76% | 8 |
| 4 | WESTRA, HJ , FRANKE, L , (2014) FROM GENOME TO FUNCTION BY STUDYING EQTLS.BIOCHIMICA ET BIOPHYSICA ACTA-MOLECULAR BASIS OF DISEASE. VOL. 1842. ISSUE 10. P. 1896 -1902 | 48 | 68% | 26 |
| 5 | ROCKMAN, MV , (2008) REVERSE ENGINEERING THE GENOTYPE-PHENOTYPE MAP WITH NATURAL GENETIC VARIATION.NATURE. VOL. 456. ISSUE 7223. P. 738 -744 | 48 | 71% | 110 |
| 6 | GIBSON, G , POWELL, JE , MARIGORTA, UM , (2015) EXPRESSION QUANTITATIVE TRAIT LOCUS ANALYSIS FOR TRANSLATIONAL MEDICINE.GENOME MEDICINE. VOL. 7. ISSUE . P. - | 58 | 55% | 6 |
| 7 | CHEUNG, VG , SPIELMAN, RS , (2009) GENETICS OF HUMAN GENE EXPRESSION: MAPPING DNA VARIANTS THAT INFLUENCE GENE EXPRESSION.NATURE REVIEWS GENETICS. VOL. 10. ISSUE 9. P. 595 -604 | 43 | 61% | 128 |
| 8 | CIVELEK, M , LUSIS, AJ , (2014) SYSTEMS GENETICS APPROACHES TO UNDERSTAND COMPLEX TRAITS.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 1. P. 34-48 | 45 | 38% | 110 |
| 9 | YANG, X , (2012) USE OF FUNCTIONAL GENOMICS TO IDENTIFY CANDIDATE GENES UNDERLYING HUMAN GENETIC ASSOCIATION STUDIES OF VASCULAR DISEASES.ARTERIOSCLEROSIS THROMBOSIS AND VASCULAR BIOLOGY. VOL. 32. ISSUE 2. P. 216-222 | 39 | 71% | 7 |
| 10 | RAKITSCH, B , STEGLE, O , (2016) MODELLING LOCAL GENE NETWORKS INCREASES POWER TO DETECT TRANS-ACTING GENETIC EFFECTS ON GENE EXPRESSION.GENOME BIOLOGY. VOL. 17. ISSUE . P. - | 40 | 65% | 0 |
Classes with closest relation at Level 1 |