Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
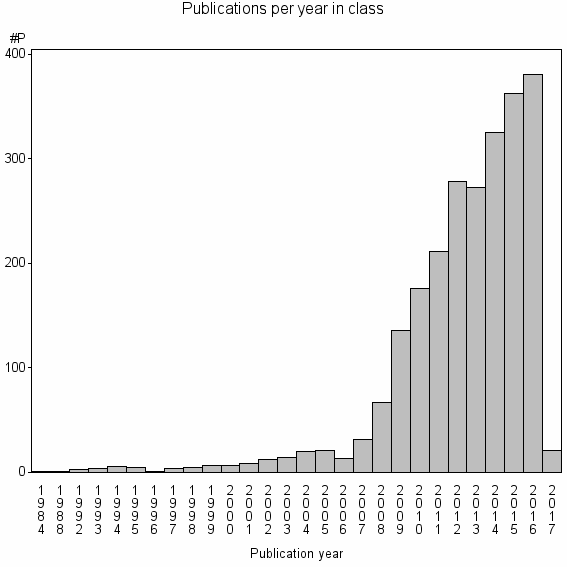
| 2013 | 2386 | 32.7 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 1101 | 2 | BIOINFORMATICS//COPY NUMBER VARIATION//NEXT GENERATION SEQUENCING | 9808 |
| 2013 | 1 | GENOME ASSEMBLY//BIOINFORMATICS//VARIANT CALLING | 2386 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENOME ASSEMBLY | authKW | 247821 | 2% | 37% | 53 |
| 2 | BIOINFORMATICS | journal | 209211 | 17% | 4% | 417 |
| 3 | VARIANT CALLING | authKW | 178114 | 1% | 61% | 23 |
| 4 | NEXT GENERATION SEQUENCING | authKW | 163766 | 10% | 5% | 236 |
| 5 | MATHEMATICAL & COMPUTATIONAL BIOLOGY | WoSSC | 104985 | 38% | 1% | 917 |
| 6 | READ MAPPING | authKW | 98426 | 0% | 77% | 10 |
| 7 | SIMONS QUANTITAT BIOL | address | 96329 | 1% | 47% | 16 |
| 8 | GIGASCIENCE | journal | 78831 | 1% | 21% | 30 |
| 9 | BMC BIOINFORMATICS | journal | 77240 | 9% | 3% | 213 |
| 10 | NANOPORE SEQUENCING | authKW | 76763 | 1% | 50% | 12 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 104985 | 38% | 1% | 917 |
| 2 | Biotechnology & Applied Microbiology | 46374 | 58% | 0% | 1394 |
| 3 | Biochemical Research Methods | 41555 | 44% | 0% | 1046 |
| 4 | Genetics & Heredity | 13333 | 32% | 0% | 752 |
| 5 | Biochemistry & Molecular Biology | 881 | 20% | 0% | 472 |
| 6 | Computer Science, Interdisciplinary Applications | 358 | 4% | 0% | 94 |
| 7 | Statistics & Probability | 230 | 3% | 0% | 70 |
| 8 | Computer Science, Theory & Methods | 90 | 2% | 0% | 57 |
| 9 | Multidisciplinary Sciences | 69 | 1% | 0% | 28 |
| 10 | Medical Informatics | 58 | 1% | 0% | 18 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SIMONS QUANTITAT BIOL | 96329 | 1% | 47% | 16 |
| 2 | STANLEY COGNIT GENOM | 30451 | 0% | 24% | 10 |
| 3 | HKU BGI BIOINFORMAT ALGORITHMS | 28789 | 0% | 75% | 3 |
| 4 | GRYC I3M | 25592 | 0% | 100% | 2 |
| 5 | INFORMAT COMP CIR | 25592 | 0% | 100% | 2 |
| 6 | INVEST SISTEMAS INTELIGENTES | 25592 | 0% | 100% | 2 |
| 7 | TEXAS ANIM HLTH COMMISS | 25592 | 0% | 100% | 2 |
| 8 | HKU BGI BIOINFORMAT ALGORITHMS CORE TECHNOL | 24601 | 0% | 38% | 5 |
| 9 | STANFORD GENOME TECHNOL | 20002 | 1% | 7% | 22 |
| 10 | ALGORITHM BIOL | 19917 | 0% | 17% | 9 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 209211 | 17% | 4% | 417 |
| 2 | GIGASCIENCE | 78831 | 1% | 21% | 30 |
| 3 | BMC BIOINFORMATICS | 77240 | 9% | 3% | 213 |
| 4 | GENOME RESEARCH | 25082 | 2% | 4% | 52 |
| 5 | BMC GENOMICS | 22465 | 5% | 1% | 130 |
| 6 | BRIEFINGS IN BIOINFORMATICS | 18792 | 1% | 5% | 32 |
| 7 | GENOME BIOLOGY | 16401 | 3% | 2% | 62 |
| 8 | JOURNAL OF COMPUTATIONAL BIOLOGY | 11793 | 2% | 2% | 38 |
| 9 | NATURE METHODS | 11440 | 2% | 2% | 39 |
| 10 | BIOTECHFORUM ( BTF ) | 9079 | 2% | 2% | 43 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENOME ASSEMBLY | 247821 | 2% | 37% | 53 | Search GENOME+ASSEMBLY | Search GENOME+ASSEMBLY |
| 2 | VARIANT CALLING | 178114 | 1% | 61% | 23 | Search VARIANT+CALLING | Search VARIANT+CALLING |
| 3 | NEXT GENERATION SEQUENCING | 163766 | 10% | 5% | 236 | Search NEXT+GENERATION+SEQUENCING | Search NEXT+GENERATION+SEQUENCING |
| 4 | READ MAPPING | 98426 | 0% | 77% | 10 | Search READ+MAPPING | Search READ+MAPPING |
| 5 | NANOPORE SEQUENCING | 76763 | 1% | 50% | 12 | Search NANOPORE+SEQUENCING | Search NANOPORE+SEQUENCING |
| 6 | MINION | 63980 | 0% | 100% | 5 | Search MINION | Search MINION |
| 7 | SHORT READ ALIGNMENT | 53315 | 0% | 83% | 5 | Search SHORT+READ+ALIGNMENT | Search SHORT+READ+ALIGNMENT |
| 8 | DE BRUIJN GRAPH | 52735 | 1% | 21% | 20 | Search DE+BRUIJN+GRAPH | Search DE+BRUIJN+GRAPH |
| 9 | OXFORD NANOPORE | 51184 | 0% | 100% | 4 | Search OXFORD+NANOPORE | Search OXFORD+NANOPORE |
| 10 | GENOME FINISHING | 45697 | 0% | 71% | 5 | Search GENOME+FINISHING | Search GENOME+FINISHING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LAEHNEMANN, D , BORKHARDT, A , MCHARDY, AC , (2016) DENOISING DNA DEEP SEQUENCING DATA-HIGH-THROUGHPUT SEQUENCING ERRORS AND THEIR CORRECTION.BRIEFINGS IN BIOINFORMATICS. VOL. 17. ISSUE 1. P. 154 -179 | 78 | 90% | 16 |
| 2 | EL-METWALLY, S , HAMZA, T , ZAKARIA, M , HELMY, M , (2013) NEXT-GENERATION SEQUENCE ASSEMBLY: FOUR STAGES OF DATA PROCESSING AND COMPUTATIONAL CHALLENGES.PLOS COMPUTATIONAL BIOLOGY. VOL. 9. ISSUE 12. P. - | 79 | 86% | 29 |
| 3 | MILLER, JR , KOREN, S , SUTTON, G , (2010) ASSEMBLY ALGORITHMS FOR NEXT-GENERATION SEQUENCING DATA.GENOMICS. VOL. 95. ISSUE 6. P. 315 -327 | 58 | 81% | 315 |
| 4 | METZKER, ML , (2010) APPLICATIONS OF NEXT-GENERATION SEQUENCING SEQUENCING TECHNOLOGIES - THE NEXT GENERATION.NATURE REVIEWS GENETICS. VOL. 11. ISSUE 1. P. 31 -46 | 42 | 46% | 2273 |
| 5 | HE, YM , ZHANG, Z , PENG, XQ , WU, FX , WANG, JX , (2013) DE NOVO ASSEMBLY METHODS FOR NEXT GENERATION SEQUENCING DATA.TSINGHUA SCIENCE AND TECHNOLOGY. VOL. 18. ISSUE 5. P. 500 -514 | 69 | 90% | 4 |
| 6 | ALTSHULER, D , DALY, MJ , DEPRISTO, MA , BANKS, E , POPLIN, R , GARIMELLA, KV , MAGUIRE, JR , HARTL, C , PHILIPPAKIS, AA , DEL ANGEL, G , ET AL (2011) A FRAMEWORK FOR VARIATION DISCOVERY AND GENOTYPING USING NEXT-GENERATION DNA SEQUENCING DATA.NATURE GENETICS. VOL. 43. ISSUE 5. P. 491 -+ | 23 | 66% | 2297 |
| 7 | LI, H , DURBIN, R , (2009) FAST AND ACCURATE SHORT READ ALIGNMENT WITH BURROWS-WHEELER TRANSFORM.BIOINFORMATICS. VOL. 25. ISSUE 14. P. 1754 -1760 | 12 | 86% | 6933 |
| 8 | PASZKIEWICZ, K , STUDHOLME, DJ , (2010) DE NOVO ASSEMBLY OF SHORT SEQUENCE READS.BRIEFINGS IN BIOINFORMATICS. VOL. 11. ISSUE 5. P. 457-472 | 65 | 78% | 65 |
| 9 | SIMPSON, JT , POP, M , (2015) THE THEORY AND PRACTICE OF GENOME SEQUENCE ASSEMBLY.ANNUAL REVIEW OF GENOMICS AND HUMAN GENETICS, VOL 16. VOL. 16. ISSUE . P. 153 -172 | 68 | 72% | 6 |
| 10 | ALIC, AS , RUZAFA, D , DOPAZO, J , BLANQUER, I , (2016) OBJECTIVE REVIEW OF DE NOVO STAND-ALONE ERROR CORRECTION METHODS FOR NGS DATA.WILEY INTERDISCIPLINARY REVIEWS-COMPUTATIONAL MOLECULAR SCIENCE. VOL. 6. ISSUE 2. P. 111 -146 | 69 | 59% | 2 |
Classes with closest relation at Level 1 |