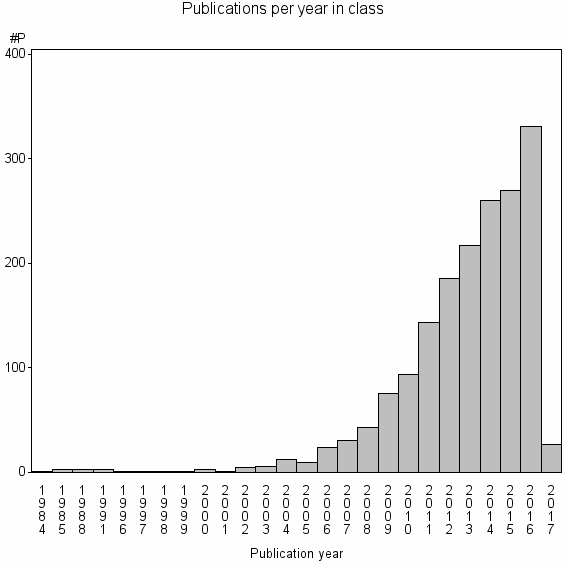
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4702 | 1741 | 50.6 | 91% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 343 | 2 | EZH2//CHROMATIN//POLYCOMB | 17754 |
| 4702 | 1 | CHIP SEQ//ENHANCER//PEAK CALLING | 1741 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CHIP SEQ | authKW | 451430 | 6% | 25% | 103 |
| 2 | ENHANCER | authKW | 108902 | 6% | 6% | 98 |
| 3 | PEAK CALLING | authKW | 95477 | 0% | 78% | 7 |
| 4 | VERTEBRATE GENOM GRP | address | 87686 | 0% | 100% | 5 |
| 5 | GENOME RESEARCH | journal | 85669 | 5% | 6% | 82 |
| 6 | ENHANCER RNA | authKW | 73070 | 0% | 83% | 5 |
| 7 | MASSIVELY PARALLEL REPORTER ASSAYS | authKW | 70149 | 0% | 100% | 4 |
| 8 | PROGRAM COMPUTAT BIOL BIOINFORMAT | address | 59360 | 2% | 9% | 37 |
| 9 | DNASE SEQ | authKW | 57389 | 0% | 55% | 6 |
| 10 | CHROMATIN LANDSCAPE | authKW | 56118 | 0% | 80% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 14767 | 17% | 0% | 297 |
| 2 | Genetics & Heredity | 12490 | 35% | 0% | 618 |
| 3 | Biotechnology & Applied Microbiology | 10523 | 33% | 0% | 580 |
| 4 | Biochemical Research Methods | 7754 | 23% | 0% | 395 |
| 5 | Biochemistry & Molecular Biology | 4280 | 42% | 0% | 737 |
| 6 | Cell Biology | 3104 | 23% | 0% | 397 |
| 7 | Developmental Biology | 200 | 3% | 0% | 44 |
| 8 | Multidisciplinary Sciences | 120 | 2% | 0% | 29 |
| 9 | Biology | 85 | 3% | 0% | 47 |
| 10 | Cell & Tissue Engineering | 60 | 1% | 0% | 11 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | VERTEBRATE GENOM GRP | 87686 | 0% | 100% | 5 |
| 2 | PROGRAM COMPUTAT BIOL BIOINFORMAT | 59360 | 2% | 9% | 37 |
| 3 | COLUMBIA STEM CELL INITIATMED | 35074 | 0% | 100% | 2 |
| 4 | GROWTH CONTROL CANC | 35074 | 0% | 100% | 2 |
| 5 | PRINCESS MARGARET CANC CENTRE | 35074 | 0% | 100% | 2 |
| 6 | FUNCT CANC EPIGENET | 31547 | 1% | 15% | 12 |
| 7 | GENOME TECHNOL BIOL GRP | 28689 | 0% | 27% | 6 |
| 8 | QUANTITAT BIOMED SCI | 24737 | 1% | 12% | 12 |
| 9 | GENOME SCI SYST BIOL | 23652 | 1% | 8% | 17 |
| 10 | MED COUNCIL MRC MOL HAEMATOL UNIT | 23382 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOME RESEARCH | 85669 | 5% | 6% | 82 |
| 2 | EPIGENETICS & CHROMATIN | 34909 | 1% | 8% | 24 |
| 3 | BIOINFORMATICS | 27717 | 7% | 1% | 130 |
| 4 | GENOME BIOLOGY | 12921 | 3% | 2% | 47 |
| 5 | NATURE METHODS | 10563 | 2% | 2% | 32 |
| 6 | BMC GENOMICS | 10216 | 4% | 1% | 75 |
| 7 | NUCLEIC ACIDS RESEARCH | 8684 | 8% | 0% | 141 |
| 8 | NATURE REVIEWS GENETICS | 7132 | 1% | 2% | 22 |
| 9 | BMC BIOINFORMATICS | 6987 | 3% | 1% | 55 |
| 10 | CELL | 4870 | 4% | 0% | 65 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CHIP SEQ | 451430 | 6% | 25% | 103 | Search CHIP+SEQ | Search CHIP+SEQ |
| 2 | ENHANCER | 108902 | 6% | 6% | 98 | Search ENHANCER | Search ENHANCER |
| 3 | PEAK CALLING | 95477 | 0% | 78% | 7 | Search PEAK+CALLING | Search PEAK+CALLING |
| 4 | ENHANCER RNA | 73070 | 0% | 83% | 5 | Search ENHANCER+RNA | Search ENHANCER+RNA |
| 5 | MASSIVELY PARALLEL REPORTER ASSAYS | 70149 | 0% | 100% | 4 | Search MASSIVELY+PARALLEL+REPORTER+ASSAYS | Search MASSIVELY+PARALLEL+REPORTER+ASSAYS |
| 6 | DNASE SEQ | 57389 | 0% | 55% | 6 | Search DNASE+SEQ | Search DNASE+SEQ |
| 7 | CHROMATIN LANDSCAPE | 56118 | 0% | 80% | 4 | Search CHROMATIN+LANDSCAPE | Search CHROMATIN+LANDSCAPE |
| 8 | ENHANCER PREDICTION | 56118 | 0% | 80% | 4 | Search ENHANCER+PREDICTION | Search ENHANCER+PREDICTION |
| 9 | ERNA | 56110 | 0% | 40% | 8 | Search ERNA | Search ERNA |
| 10 | HISTONE MODIFICATION | 54508 | 4% | 4% | 75 | Search HISTONE+MODIFICATION | Search HISTONE+MODIFICATION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SHLYUEVA, D , STAMPFEL, G , STARK, A , (2014) TRANSCRIPTIONAL ENHANCERS: FROM PROPERTIES TO GENOME-WIDE PREDICTIONS.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 4. P. 272 -286 | 61 | 40% | 177 |
| 2 | KLEFTOGIANNIS, D , KALNIS, P , BAJIC, VB , (2016) PROGRESS AND CHALLENGES IN BIOINFORMATICS APPROACHES FOR ENHANCER IDENTIFICATION.BRIEFINGS IN BIOINFORMATICS. VOL. 17. ISSUE 6. P. 967 -979 | 69 | 72% | 0 |
| 3 | ANDERSSON, R , (2015) PROMOTER OR ENHANCER, WHAT'S THE DIFFERENCE? DECONSTRUCTION OF ESTABLISHED DISTINCTIONS AND PRESENTATION OF A UNIFYING MODEL.BIOESSAYS. VOL. 37. ISSUE 3. P. 314 -323 | 67 | 58% | 14 |
| 4 | BAILEY, T , KRAJEWSKI, P , LADUNGA, I , LEFEBVRE, C , LI, QH , LIU, T , MADRIGAL, P , TASLIM, C , ZHANG, J , (2013) PRACTICAL GUIDELINES FOR THE COMPREHENSIVE ANALYSIS OF CHIP-SEQ DATA.PLOS COMPUTATIONAL BIOLOGY. VOL. 9. ISSUE 11. P. - | 59 | 66% | 47 |
| 5 | LI, WB , NOTANI, D , ROSENFELD, MG , (2016) ENHANCERS AS NON-CODING RNA TRANSCRIPTION UNITS: RECENT INSIGHTS AND FUTURE PERSPECTIVES.NATURE REVIEWS GENETICS. VOL. 17. ISSUE 4. P. 207 -223 | 62 | 35% | 15 |
| 6 | HEINZ, S , ROMANOSKI, CE , BENNER, C , GLASS, CK , (2015) THE SELECTION AND FUNCTION OF CELL TYPE-SPECIFIC ENHANCERS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 16. ISSUE 3. P. 144 -154 | 49 | 39% | 69 |
| 7 | MEYER, CA , LIU, XS , (2014) IDENTIFYING AND MITIGATING BIAS IN NEXT-GENERATION SEQUENCING METHODS FOR CHROMATIN BIOLOGY.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 11. P. 709 -721 | 54 | 50% | 34 |
| 8 | KUNDAJE, A , MEULEMAN, W , ERNST, J , BILENKY, M , YEN, A , HERAVI-MOUSSAVI, A , KHERADPOUR, P , ZHANG, Z , WANG, J , ZILLER, MJ , ET AL (2015) INTEGRATIVE ANALYSIS OF 111 REFERENCE HUMAN EPIGENOMES.NATURE. VOL. 518. ISSUE 7539. P. 317 -330 | 32 | 32% | 238 |
| 9 | YAN, LY , GUO, HS , HU, BQ , LI, R , YONG, J , ZHAO, YY , ZHI, X , FAN, XY , GUO, F , WANG, XY , ET AL (2016) EPIGENOMIC LANDSCAPE OF HUMAN FETAL BRAIN, HEART, AND LIVER.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 291. ISSUE 9. P. 4386 -4398 | 38 | 76% | 1 |
| 10 | HAH, N , BENNER, C , CHONG, LW , YU, RT , DOWNES, M , EVANS, RM , (2015) INFLAMMATION-SENSITIVE SUPER ENHANCERS FORM DOMAINS OF COORDINATELY REGULATED ENHANCER RNAS.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. VOL. 112. ISSUE 3. P. E297 -E302 | 32 | 73% | 21 |
Classes with closest relation at Level 1 |