Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
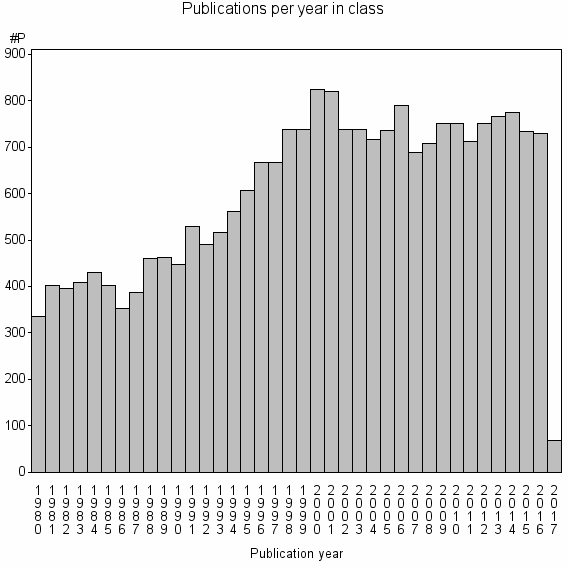
| 160 | 22798 | 44.9 | 78% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | WERNER SYNDROME | authKW | 281525 | 1% | 79% | 266 |
| 2 | DNA POLYMERASE | authKW | 245961 | 2% | 40% | 460 |
| 3 | DNA REPAIR | journal | 226065 | 3% | 29% | 587 |
| 4 | DNA REPLICATION | authKW | 197837 | 3% | 23% | 653 |
| 5 | TRANSLESION SYNTHESIS | authKW | 187161 | 1% | 77% | 181 |
| 6 | MISMATCH REPAIR | authKW | 140332 | 1% | 34% | 312 |
| 7 | HOMOLOGOUS RECOMBINATION | authKW | 137195 | 2% | 21% | 489 |
| 8 | RECA PROTEIN | authKW | 134989 | 1% | 88% | 115 |
| 9 | HOLLIDAY JUNCTION | authKW | 134897 | 1% | 65% | 155 |
| 10 | HELICASE | authKW | 121807 | 1% | 37% | 249 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 116048 | 58% | 1% | 13335 |
| 2 | Genetics & Heredity | 62786 | 23% | 1% | 5153 |
| 3 | Cell Biology | 18364 | 16% | 1% | 3670 |
| 4 | Toxicology | 9059 | 7% | 1% | 1523 |
| 5 | Microbiology | 6460 | 8% | 0% | 1913 |
| 6 | Biophysics | 4154 | 6% | 0% | 1359 |
| 7 | Biology | 1415 | 3% | 0% | 672 |
| 8 | Biotechnology & Applied Microbiology | 1139 | 5% | 0% | 1034 |
| 9 | Oncology | 584 | 5% | 0% | 1118 |
| 10 | Biochemical Research Methods | 475 | 3% | 0% | 577 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SECT DNA REPLICAT REPAIR MUTAGENESIS | 78712 | 0% | 89% | 66 |
| 2 | MOL GERONTOL | 69217 | 1% | 31% | 169 |
| 3 | GENOM INTEGR | 50494 | 0% | 79% | 48 |
| 4 | CLARE HALL S | 42585 | 1% | 22% | 147 |
| 5 | HEDCO MOL BIOL S | 32592 | 0% | 76% | 32 |
| 6 | DNA REPLICAT | 31833 | 0% | 74% | 32 |
| 7 | STRUCT BIOL | 27490 | 2% | 6% | 373 |
| 8 | SEALY MOL SCI | 24923 | 0% | 23% | 82 |
| 9 | CREAT INITIAT CELL CYCLE CONTROL | 24073 | 0% | 100% | 18 |
| 10 | BRAUN S | 18478 | 0% | 63% | 22 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 226065 | 3% | 29% | 587 |
| 2 | NUCLEIC ACIDS RESEARCH | 78298 | 7% | 4% | 1537 |
| 3 | JOURNAL OF BIOLOGICAL CHEMISTRY | 46610 | 10% | 2% | 2360 |
| 4 | GENETICS | 40025 | 3% | 5% | 625 |
| 5 | JOURNAL OF MOLECULAR BIOLOGY | 36255 | 4% | 4% | 807 |
| 6 | JOURNAL OF BACTERIOLOGY | 25655 | 4% | 2% | 816 |
| 7 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 25145 | 1% | 7% | 273 |
| 8 | MOLECULAR AND GENERAL GENETICS | 22318 | 1% | 6% | 286 |
| 9 | MUTATION RESEARCH-DNA REPAIR | 20170 | 0% | 18% | 86 |
| 10 | BIOCHEMISTRY | 19758 | 4% | 2% | 917 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | WERNER SYNDROME | 281525 | 1% | 79% | 266 | Search WERNER+SYNDROME | Search WERNER+SYNDROME |
| 2 | DNA POLYMERASE | 245961 | 2% | 40% | 460 | Search DNA+POLYMERASE | Search DNA+POLYMERASE |
| 3 | DNA REPLICATION | 197837 | 3% | 23% | 653 | Search DNA+REPLICATION | Search DNA+REPLICATION |
| 4 | TRANSLESION SYNTHESIS | 187161 | 1% | 77% | 181 | Search TRANSLESION+SYNTHESIS | Search TRANSLESION+SYNTHESIS |
| 5 | MISMATCH REPAIR | 140332 | 1% | 34% | 312 | Search MISMATCH+REPAIR | Search MISMATCH+REPAIR |
| 6 | DNA REPAIR | 140046 | 4% | 12% | 865 | Search DNA+REPAIR | Search DNA+REPAIR |
| 7 | HOMOLOGOUS RECOMBINATION | 137195 | 2% | 21% | 489 | Search HOMOLOGOUS+RECOMBINATION | Search HOMOLOGOUS+RECOMBINATION |
| 8 | RECA PROTEIN | 134989 | 1% | 88% | 115 | Search RECA+PROTEIN | Search RECA+PROTEIN |
| 9 | HOLLIDAY JUNCTION | 134897 | 1% | 65% | 155 | Search HOLLIDAY+JUNCTION | Search HOLLIDAY+JUNCTION |
| 10 | HELICASE | 121807 | 1% | 37% | 249 | Search HELICASE | Search HELICASE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | KOWALCZYKOWSKI, SC , DIXON, DA , EGGLESTON, AK , LAUDER, SD , REHRAUER, WM , (1994) BIOCHEMISTRY OF HOMOLOGOUS RECOMBINATION IN ESCHERICHIA-COLI.MICROBIOLOGICAL REVIEWS. VOL. 58. ISSUE 3. P. 401 -465 | 507 | 91% | 928 |
| 2 | KUZMINOV, A , (1999) RECOMBINATIONAL REPAIR OF DNA DAMAGE IN ESCHERICHIA COLI AND BACTERIOPHAGE LAMBDA.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 63. ISSUE 4. P. 751 -+ | 442 | 80% | 584 |
| 3 | SHEREDA, RD , KOZLOV, AG , LOHMAN, TM , COX, MM , KECK, JL , (2008) SSB AS AN ORGANIZER/MOBILIZER OF GENOME MAINTENANCE COMPLEXES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 43. ISSUE 5. P. 289 -318 | 352 | 92% | 161 |
| 4 | KOWALCZYKOWSKI, SC , (2015) AN OVERVIEW OF THE MOLECULAR MECHANISMS OF RECOMBINATIONAL DNA REPAIR.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 7. ISSUE 11. P. - | 287 | 80% | 21 |
| 5 | IYER, RR , PLUCIENNIK, A , BURDETT, V , MODRICH, PL , (2006) DNA MISMATCH REPAIR: FUNCTIONS AND MECHANISMS.CHEMICAL REVIEWS. VOL. 106. ISSUE 2. P. 302 -323 | 293 | 79% | 416 |
| 6 | CROTEAU, DL , POPURI, V , OPRESKO, PL , BOHR, VA , (2014) HUMAN RECQ HELICASES IN DNA REPAIR, RECOMBINATION, AND REPLICATION.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 83. VOL. 83. ISSUE . P. 519 -552 | 222 | 76% | 79 |
| 7 | WATERS, LS , MINESINGER, BK , WILTROUT, ME , D'SOUZA, S , WOODRUFF, RV , WALKER, GC , (2009) EUKARYOTIC TRANSLESION POLYMERASES AND THEIR ROLES AND REGULATION IN DNA DAMAGE TOLERANCE.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 73. ISSUE 1. P. 134-+ | 229 | 85% | 241 |
| 8 | COX, MM , (2007) REGULATION OF BACTERIAL RECA PROTEIN FUNCTION.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 42. ISSUE 1. P. 41 -63 | 233 | 90% | 161 |
| 9 | DILLINGHAM, MS , KOWALCZYKOWSKI, SC , (2008) RECBCD ENZYME AND THE REPAIR OF DOUBLE-STRANDED DNA BREAKS.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 72. ISSUE 4. P. 642-+ | 218 | 87% | 172 |
| 10 | BICHARA, M , MEIER, M , WAGNER, J , CORDONNIER, A , LAMBERT, IB , (2011) POSTREPLICATION REPAIR MECHANISMS IN THE PRESENCE OF DNA ADDUCTS IN ESCHERICHIA COLI.MUTATION RESEARCH-REVIEWS IN MUTATION RESEARCH. VOL. 727. ISSUE 3. P. 104-122 | 241 | 95% | 10 |
Classes with closest relation at Level 2 |