Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
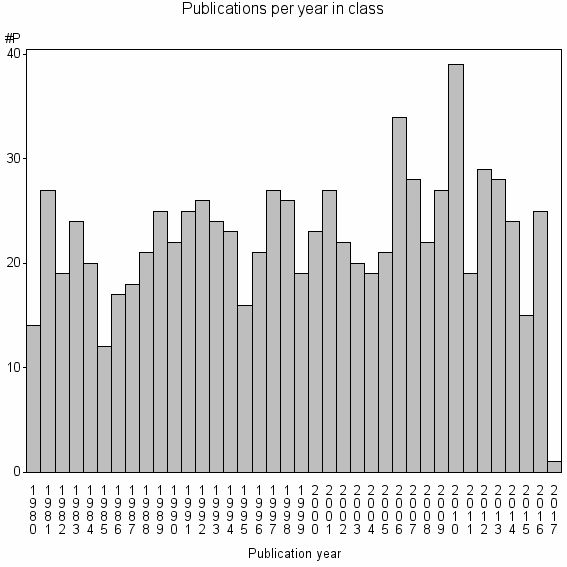
| 13276 | 849 | 43.0 | 77% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 13276 | 1 | UVRB//UVRD//UVRABC | 849 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | UVRB | authKW | 575429 | 2% | 80% | 20 |
| 2 | UVRD | authKW | 235395 | 1% | 55% | 12 |
| 3 | UVRABC | authKW | 224083 | 1% | 69% | 9 |
| 4 | HELICASE | authKW | 184272 | 7% | 9% | 59 |
| 5 | UVRA | authKW | 153314 | 1% | 47% | 9 |
| 6 | DIMER EXCISION | authKW | 143859 | 0% | 100% | 4 |
| 7 | HELICASE II | authKW | 143859 | 0% | 100% | 4 |
| 8 | ABC EXCINUCLEASE | authKW | 107894 | 0% | 100% | 3 |
| 9 | PCRA | authKW | 103655 | 1% | 41% | 7 |
| 10 | UVRC | authKW | 99898 | 1% | 56% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 7191 | 74% | 0% | 627 |
| 2 | Genetics & Heredity | 684 | 13% | 0% | 110 |
| 3 | Biophysics | 464 | 9% | 0% | 80 |
| 4 | Toxicology | 314 | 6% | 0% | 55 |
| 5 | Microbiology | 300 | 9% | 0% | 78 |
| 6 | Cell Biology | 283 | 11% | 0% | 95 |
| 7 | Biology | 78 | 3% | 0% | 29 |
| 8 | Biochemical Research Methods | 33 | 3% | 0% | 26 |
| 9 | Mathematical & Computational Biology | 4 | 1% | 0% | 6 |
| 10 | Biotechnology & Applied Microbiology | 1 | 2% | 0% | 17 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA PROT INTERACT UNIT | 84906 | 2% | 17% | 14 |
| 2 | PROT ENGN MOL GENET TRAINING PROGRAM | 71930 | 0% | 100% | 2 |
| 3 | ZELLULARE PHYSIOL | 71930 | 0% | 100% | 2 |
| 4 | BIOCHIM CHIM PROT | 47952 | 0% | 67% | 2 |
| 5 | BIOL PHARMACOL PLIDA FR3242 | 35965 | 0% | 100% | 1 |
| 6 | BIOSPECT CORE | 35965 | 0% | 100% | 1 |
| 7 | BIOTECHNOL INFORMAT LIBRARY MED | 35965 | 0% | 100% | 1 |
| 8 | BLDG 38A8600 ROCKVILLE PIKE | 35965 | 0% | 100% | 1 |
| 9 | C3MEQUIPE CONTROLE METABOL MORTS CELLULAI | 35965 | 0% | 100% | 1 |
| 10 | CNRSIBENS | 35965 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 10152 | 3% | 1% | 24 |
| 2 | NUCLEIC ACIDS RESEARCH | 9047 | 12% | 0% | 100 |
| 3 | JOURNAL OF BIOLOGICAL CHEMISTRY | 3613 | 15% | 0% | 125 |
| 4 | JOURNAL OF MOLECULAR BIOLOGY | 3211 | 5% | 0% | 46 |
| 5 | BIOCHEMISTRY | 2201 | 7% | 0% | 58 |
| 6 | JOURNAL OF BACTERIOLOGY | 2041 | 5% | 0% | 44 |
| 7 | MUTATION RESEARCH-DNA REPAIR | 1836 | 1% | 1% | 5 |
| 8 | MUTATION RESEARCH | 1571 | 2% | 0% | 19 |
| 9 | EMBO JOURNAL | 984 | 3% | 0% | 22 |
| 10 | METHODS | 944 | 1% | 0% | 9 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | UVRB | 575429 | 2% | 80% | 20 | Search UVRB | Search UVRB |
| 2 | UVRD | 235395 | 1% | 55% | 12 | Search UVRD | Search UVRD |
| 3 | UVRABC | 224083 | 1% | 69% | 9 | Search UVRABC | Search UVRABC |
| 4 | HELICASE | 184272 | 7% | 9% | 59 | Search HELICASE | Search HELICASE |
| 5 | UVRA | 153314 | 1% | 47% | 9 | Search UVRA | Search UVRA |
| 6 | DIMER EXCISION | 143859 | 0% | 100% | 4 | Search DIMER+EXCISION | Search DIMER+EXCISION |
| 7 | HELICASE II | 143859 | 0% | 100% | 4 | Search HELICASE+II | Search HELICASE+II |
| 8 | ABC EXCINUCLEASE | 107894 | 0% | 100% | 3 | Search ABC+EXCINUCLEASE | Search ABC+EXCINUCLEASE |
| 9 | PCRA | 103655 | 1% | 41% | 7 | Search PCRA | Search PCRA |
| 10 | UVRC | 99898 | 1% | 56% | 5 | Search UVRC | Search UVRC |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | KISKER, C , KUPER, J , VAN HOUTEN, B , (2013) PROKARYOTIC NUCLEOTIDE EXCISION REPAIR.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 5. ISSUE 3. P. - | 88 | 81% | 29 |
| 2 | TRUGLIO, JJ , CROTEAU, DL , VAN HOUTEN, B , KISKER, C , (2006) PROKARYOTIC NUCLEOTIDE EXCISION REPAIR: THE UVRABC SYSTEM.CHEMICAL REVIEWS. VOL. 106. ISSUE 2. P. 233-252 | 121 | 58% | 162 |
| 3 | THAKUR, M , KUMAR, MBJ , MUNIYAPPA, K , (2016) MYCOBACTERIUM TUBERCULOSIS UVRB IS A ROBUST DNA-STIMULATED ATPASE THAT ALSO POSSESSES STRUCTURE-SPECIFIC ATP-DEPENDENT DNA HELICASE ACTIVITY.BIOCHEMISTRY. VOL. 55. ISSUE 41. P. 5865 -5883 | 65 | 88% | 0 |
| 4 | VAN HOUTEN, B , CROTEAU, DL , DELLAVECCHIA, MJ , WANG, H , KISKER, C , (2005) 'CLOSE-FITTING SLEEVES': DNA DAMAGE RECOGNITION BY THE UVRABC NUCLEASE SYSTEM.MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS. VOL. 577. ISSUE 1-2. P. 92-117 | 94 | 56% | 79 |
| 5 | DILLINGHAM, MS , (2011) SUPERFAMILY I HELICASES AS MODULAR COMPONENTS OF DNA-PROCESSING MACHINES.BIOCHEMICAL SOCIETY TRANSACTIONS. VOL. 39. ISSUE . P. 413 -423 | 67 | 60% | 23 |
| 6 | LOHMAN, TM , TOMKO, EJ , WU, CG , (2008) NON-HEXAMERIC DNA HELICASES AND TRANSLOCASES: MECHANISMS AND REGULATION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 9. ISSUE 5. P. 391 -401 | 59 | 53% | 146 |
| 7 | VAN HOUTEN, B , KAD, N , (2014) INVESTIGATION OF BACTERIAL NUCLEOTIDE EXCISION REPAIR USING SINGLE-MOLECULE TECHNIQUES.DNA REPAIR. VOL. 20. ISSUE . P. 41 -48 | 43 | 81% | 1 |
| 8 | WANG, H , DELLAVECCHIA, MJ , SKORVAGA, M , CROTEAU, DL , ERIE, DA , VAN HOUTEN, B , (2006) UVRB DOMAIN 4, AN AUTOINHIBITORY GATE FOR REGULATION OF DNA BINDING AND ATPASE ACTIVITY.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 281. ISSUE 22. P. 15227-15237 | 42 | 86% | 15 |
| 9 | COMSTOCK, MJ , WHITLEY, KD , JIA, HF , SOKOLOSKI, J , LOHMAN, TM , HA, T , CHEMLA, YR , (2015) DIRECT OBSERVATION OF STRUCTURE-FUNCTION RELATIONSHIP IN A NUCLEIC ACID-PROCESSING ENZYME.SCIENCE. VOL. 348. ISSUE 6232. P. 352 -354 | 25 | 86% | 22 |
| 10 | CROTEAU, DL , DELLAVECCHIA, MJ , PERERA, L , VAN HOUTEN, B , (2008) COOPERATIVE DAMAGE RECOGNITION BY UVRA AND UVRB: IDENTIFICATION OF UVRA RESIDUES THAT MEDIATE DNA BINDING.DNA REPAIR. VOL. 7. ISSUE 3. P. 392-404 | 39 | 83% | 16 |
Classes with closest relation at Level 1 |