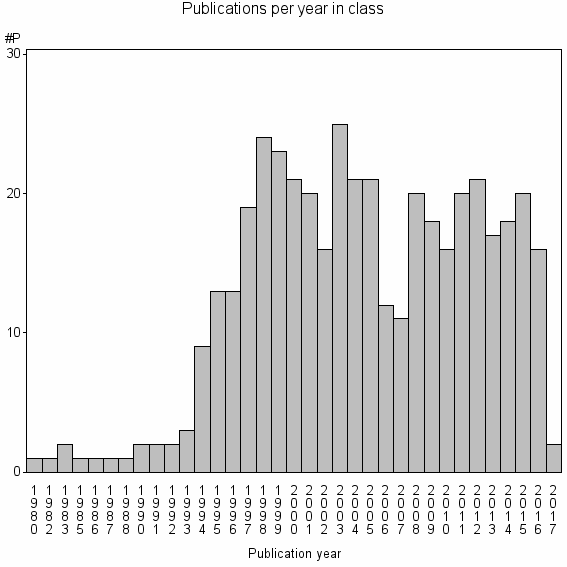
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 21108 | 433 | 46.3 | 89% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 21108 | 1 | FEN1//BRAUN S//CREAT INITIAT CELL CYCLE CONTROL | 433 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | FEN1 | authKW | 1220505 | 7% | 58% | 30 |
| 2 | BRAUN S | address | 805920 | 5% | 57% | 20 |
| 3 | CREAT INITIAT CELL CYCLE CONTROL | address | 662093 | 3% | 72% | 13 |
| 4 | FLAP ENDONUCLEASE 1 | authKW | 634664 | 3% | 60% | 15 |
| 5 | OKAZAKI FRAGMENT PROCESSING | authKW | 451322 | 2% | 80% | 8 |
| 6 | DNA2 | authKW | 441502 | 3% | 52% | 12 |
| 7 | FEN 1 | authKW | 414813 | 2% | 59% | 10 |
| 8 | FLAP ENDONUCLEASE | authKW | 414813 | 2% | 59% | 10 |
| 9 | RAD27 | authKW | 345542 | 2% | 70% | 7 |
| 10 | DNA REPLICAT GENOME ABIL | address | 282074 | 1% | 67% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 3673 | 74% | 0% | 320 |
| 2 | Cell Biology | 627 | 21% | 0% | 90 |
| 3 | Genetics & Heredity | 319 | 12% | 0% | 54 |
| 4 | Toxicology | 63 | 4% | 0% | 19 |
| 5 | Biophysics | 40 | 5% | 0% | 20 |
| 6 | Oncology | 40 | 7% | 0% | 31 |
| 7 | Biochemical Research Methods | 20 | 3% | 0% | 14 |
| 8 | Biotechnology & Applied Microbiology | 13 | 4% | 0% | 17 |
| 9 | Biology | 10 | 2% | 0% | 9 |
| 10 | Crystallography | 1 | 1% | 0% | 5 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BRAUN S | 805920 | 5% | 57% | 20 |
| 2 | CREAT INITIAT CELL CYCLE CONTROL | 662093 | 3% | 72% | 13 |
| 3 | DNA REPLICAT GENOME ABIL | 282074 | 1% | 67% | 6 |
| 4 | KREBS | 104979 | 5% | 7% | 22 |
| 5 | CELL TUMOR BIOL | 103688 | 2% | 15% | 10 |
| 6 | BRAUN S 147 75 | 102569 | 1% | 36% | 4 |
| 7 | SAMSUNG BIOMED CHANGAN KU | 94025 | 0% | 67% | 2 |
| 8 | HENRY WELLCOME S MED | 84614 | 1% | 20% | 6 |
| 9 | AIR POLLUT HLTH MED | 70520 | 0% | 100% | 1 |
| 10 | BRAUN 14775 | 70520 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BIOLOGICAL CHEMISTRY | 5231 | 24% | 0% | 106 |
| 2 | NUCLEIC ACIDS RESEARCH | 3911 | 11% | 0% | 47 |
| 3 | DNA REPAIR | 1238 | 1% | 0% | 6 |
| 4 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 1146 | 2% | 0% | 8 |
| 5 | JOURNAL OF MOLECULAR CELL BIOLOGY | 1060 | 0% | 1% | 2 |
| 6 | MOLECULAR AND CELLULAR BIOLOGY | 789 | 4% | 0% | 16 |
| 7 | MUTATION RESEARCH-DNA REPAIR | 575 | 0% | 0% | 2 |
| 8 | CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY | 530 | 0% | 0% | 2 |
| 9 | ARCHAEA-AN INTERNATIONAL MICROBIOLOGICAL JOURNAL | 474 | 0% | 1% | 1 |
| 10 | EMBO JOURNAL | 397 | 2% | 0% | 10 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | FEN1 | 1220505 | 7% | 58% | 30 | Search FEN1 | Search FEN1 |
| 2 | FLAP ENDONUCLEASE 1 | 634664 | 3% | 60% | 15 | Search FLAP+ENDONUCLEASE+1 | Search FLAP+ENDONUCLEASE+1 |
| 3 | OKAZAKI FRAGMENT PROCESSING | 451322 | 2% | 80% | 8 | Search OKAZAKI+FRAGMENT+PROCESSING | Search OKAZAKI+FRAGMENT+PROCESSING |
| 4 | DNA2 | 441502 | 3% | 52% | 12 | Search DNA2 | Search DNA2 |
| 5 | FEN 1 | 414813 | 2% | 59% | 10 | Search FEN+1 | Search FEN+1 |
| 6 | FLAP ENDONUCLEASE | 414813 | 2% | 59% | 10 | Search FLAP+ENDONUCLEASE | Search FLAP+ENDONUCLEASE |
| 7 | RAD27 | 345542 | 2% | 70% | 7 | Search RAD27 | Search RAD27 |
| 8 | OKAZAKI FRAGMENT MATURATION | 282074 | 1% | 67% | 6 | Search OKAZAKI+FRAGMENT+MATURATION | Search OKAZAKI+FRAGMENT+MATURATION |
| 9 | DNASE IV | 211559 | 1% | 100% | 3 | Search DNASE+IV | Search DNASE+IV |
| 10 | FLAP ENDONUCLEASE 1 FEN1 | 176294 | 1% | 50% | 5 | Search FLAP+ENDONUCLEASE+1+FEN1 | Search FLAP+ENDONUCLEASE+1+FEN1 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LIU, Y , KAO, HI , BAMBARA, RA , (2004) FLAP ENDONUCLEASE 1: A CENTRAL COMPONENT OF DNA METABOLISM.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 73. ISSUE . P. 589-615 | 90 | 51% | 213 |
| 2 | SHEN, BH , SINGH, P , LIU, R , QIU, JZ , ZHENG, L , FINGER, LD , ALAS, S , (2005) MULTIPLE BUT DISSECTIBLE FUNCTIONS OF FEN-1 NUCLEASES IN NUCLEIC ACID PROCESSING, GENOME STABILITY AND DISEASES.BIOESSAYS. VOL. 27. ISSUE 7. P. 717-729 | 70 | 74% | 79 |
| 3 | ZHENG, L , JIA, J , FINGER, LD , GUO, ZG , ZER, C , SHEN, BH , (2011) FUNCTIONAL REGULATION OF FEN1 NUCLEASE AND ITS LINK TO CANCER.NUCLEIC ACIDS RESEARCH. VOL. 39. ISSUE 3. P. 781-794 | 67 | 58% | 42 |
| 4 | LEVIKOVA, M , CEJKA, P , (2015) THE SACCHAROMYCES CEREVISIAE DNA2 CAN FUNCTION AS A SOLE NUCLEASE IN THE PROCESSING OF OKAZAKI FRAGMENTS IN DNA REPLICATION.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE 16. P. 7888 -7897 | 48 | 74% | 3 |
| 5 | NAZARKINA, ZK , LAVRIK, OI , KHODYREVA, SN , (2008) FLAP ENDONUCLEASE 1 AND ITS ROLE IN EUKARYOTIC DNA METABOLISM.MOLECULAR BIOLOGY. VOL. 42. ISSUE 3. P. 357-370 | 59 | 68% | 1 |
| 6 | BALAKRISHNAN, L , BAMBARA, RA , (2013) FLAP ENDONUCLEASE 1.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 82. VOL. 82. ISSUE . P. 119 -138 | 57 | 48% | 42 |
| 7 | FINGER, LD , BLANCHARD, MS , THEIMER, CA , SENGEROVA, B , SINGH, P , CHAVEZ, V , LIU, F , GRASBY, JA , SHEN, BH , (2009) THE 3 '-FLAP POCKET OF HUMAN FLAP ENDONUCLEASE 1 IS CRITICAL FOR SUBSTRATE BINDING AND CATALYSIS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 284. ISSUE 33. P. 22184 -22194 | 47 | 76% | 15 |
| 8 | GLOOR, JW , BALAKRISHNAN, L , CAMPBELL, JL , BAMBARA, RA , (2012) BIOCHEMICAL ANALYSES INDICATE THAT BINDING AND CLEAVAGE SPECIFICITIES DEFINE THE ORDERED PROCESSING OF HUMAN OKAZAKI FRAGMENTS BY DNA2 AND FEN1.NUCLEIC ACIDS RESEARCH. VOL. 40. ISSUE 14. P. 6774-6786 | 39 | 72% | 10 |
| 9 | KANG, YH , LEE, CH , SEO, YS , (2010) DNA2 ON THE ROAD TO OKAZAKI FRAGMENT PROCESSING AND GENOME STABILITY IN EUKARYOTES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 45. ISSUE 2. P. 71 -96 | 74 | 37% | 43 |
| 10 | ZHENG, L , SHEN, BH , (2011) OKAZAKI FRAGMENT MATURATION: NUCLEASES TAKE CENTRE STAGE.JOURNAL OF MOLECULAR CELL BIOLOGY. VOL. 3. ISSUE 1. P. 23 -30 | 40 | 61% | 62 |
Classes with closest relation at Level 1 |