Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
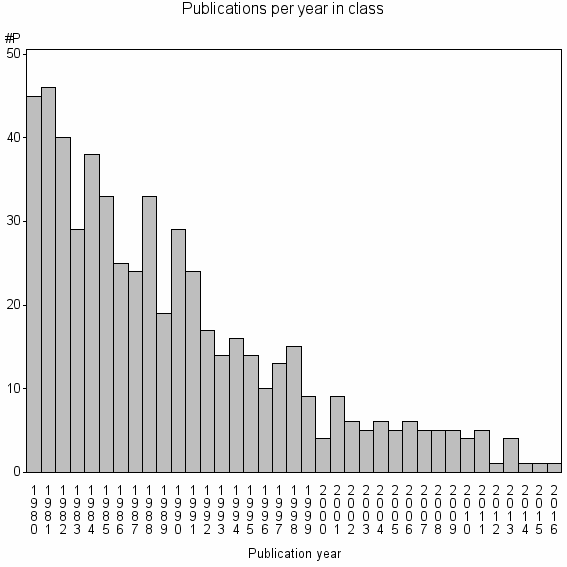
| 18097 | 566 | 40.3 | 43% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 18097 | 1 | GENE UVSX//BACTERIOPHAGE T4//UVSX | 566 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENE UVSX | authKW | 215793 | 1% | 100% | 4 |
| 2 | BACTERIOPHAGE T4 | authKW | 178546 | 4% | 14% | 24 |
| 3 | UVSX | authKW | 172633 | 1% | 80% | 4 |
| 4 | UVSY | authKW | 161845 | 1% | 100% | 3 |
| 5 | BACTERIOPHAGE SPO1 | authKW | 107896 | 0% | 100% | 2 |
| 6 | BASEPLATE GENES | authKW | 107896 | 0% | 100% | 2 |
| 7 | GENE 39 | authKW | 107896 | 0% | 100% | 2 |
| 8 | GENE 61 | authKW | 107896 | 0% | 100% | 2 |
| 9 | RECOMBINATIONAL ANALYSIS | authKW | 107896 | 0% | 100% | 2 |
| 10 | ENDONUCLEASE VII | authKW | 61649 | 1% | 29% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Virology | 3335 | 19% | 0% | 106 |
| 2 | Biochemistry & Molecular Biology | 2196 | 52% | 0% | 293 |
| 3 | Genetics & Heredity | 2191 | 27% | 0% | 150 |
| 4 | Microbiology | 191 | 9% | 0% | 51 |
| 5 | Biology | 43 | 3% | 0% | 18 |
| 6 | Toxicology | 11 | 2% | 0% | 12 |
| 7 | Biochemical Research Methods | 8 | 2% | 0% | 13 |
| 8 | Biophysics | 7 | 2% | 0% | 14 |
| 9 | Cell Biology | 7 | 4% | 0% | 23 |
| 10 | Biotechnology & Applied Microbiology | 0 | 2% | 0% | 11 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | AG MOL GENET | 53948 | 0% | 100% | 1 |
| 2 | INT MOL CELLULAR PATHOL | 53948 | 0% | 100% | 1 |
| 3 | WARTIK S 414 | 53948 | 0% | 100% | 1 |
| 4 | MOL GENET E3 01 | 43156 | 0% | 40% | 2 |
| 5 | CHEM BIOL KINET | 30825 | 0% | 29% | 2 |
| 6 | MOL GENET E301 | 26973 | 0% | 50% | 1 |
| 7 | UNIT MICROBIAL PATHOGENESIS | 26973 | 0% | 50% | 1 |
| 8 | VERMONT CANC | 21911 | 2% | 4% | 10 |
| 9 | BIOCHEM SL 43 | 13486 | 0% | 25% | 1 |
| 10 | LEHRSTUHL STRUKT CHEM BIOPOLYMERE | 13486 | 0% | 25% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENETICS | 8527 | 8% | 0% | 45 |
| 2 | MOLECULAR & GENERAL GENETICS | 7178 | 3% | 1% | 18 |
| 3 | JOURNAL OF MOLECULAR BIOLOGY | 6972 | 10% | 0% | 55 |
| 4 | VIROLOGY | 4159 | 7% | 0% | 38 |
| 5 | JOURNAL OF VIROLOGY | 3922 | 10% | 0% | 54 |
| 6 | MOLECULAR AND GENERAL GENETICS | 2506 | 3% | 0% | 15 |
| 7 | GENETIKA | 2393 | 2% | 0% | 14 |
| 8 | GENE | 1443 | 4% | 0% | 23 |
| 9 | DNA REPAIR | 1288 | 1% | 0% | 7 |
| 10 | NUCLEIC ACIDS RESEARCH | 1272 | 5% | 0% | 31 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENE UVSX | 215793 | 1% | 100% | 4 | Search GENE+UVSX | Search GENE+UVSX |
| 2 | BACTERIOPHAGE T4 | 178546 | 4% | 14% | 24 | Search BACTERIOPHAGE+T4 | Search BACTERIOPHAGE+T4 |
| 3 | UVSX | 172633 | 1% | 80% | 4 | Search UVSX | Search UVSX |
| 4 | UVSY | 161845 | 1% | 100% | 3 | Search UVSY | Search UVSY |
| 5 | BACTERIOPHAGE SPO1 | 107896 | 0% | 100% | 2 | Search BACTERIOPHAGE+SPO1 | Search BACTERIOPHAGE+SPO1 |
| 6 | BASEPLATE GENES | 107896 | 0% | 100% | 2 | Search BASEPLATE+GENES | Search BASEPLATE+GENES |
| 7 | GENE 39 | 107896 | 0% | 100% | 2 | Search GENE+39 | Search GENE+39 |
| 8 | GENE 61 | 107896 | 0% | 100% | 2 | Search GENE+61 | Search GENE+61 |
| 9 | RECOMBINATIONAL ANALYSIS | 107896 | 0% | 100% | 2 | Search RECOMBINATIONAL+ANALYSIS | Search RECOMBINATIONAL+ANALYSIS |
| 10 | ENDONUCLEASE VII | 61649 | 1% | 29% | 4 | Search ENDONUCLEASE+VII | Search ENDONUCLEASE+VII |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MILLER, ES , KUTTER, E , MOSIG, G , ARISAKA, F , KUNISAWA, T , RUGER, W , (2003) BACTERIOPHAGE T4 GENOME.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 67. ISSUE 1. P. 86 -+ | 220 | 26% | 302 |
| 2 | KREUZER, KN , BRISTER, JR , (2010) INITIATION OF BACTERIOPHAGE T4 DNA REPLICATION AND REPLICATION FORK DYNAMICS: A REVIEW IN THE VIROLOGY JOURNAL SERIES ON BACTERIOPHAGE T4 AND ITS RELATIVES.VIROLOGY JOURNAL. VOL. 7. ISSUE . P. - | 50 | 50% | 29 |
| 3 | LIU, J , MORRICAL, SW , (2010) ASSEMBLY AND DYNAMICS OF THE BACTERIOPHAGE T4 HOMOLOGOUS RECOMBINATION MACHINERY.VIROLOGY JOURNAL. VOL. 7. ISSUE . P. - | 45 | 51% | 13 |
| 4 | MOSIG, G , (1998) RECOMBINATION AND RECOMBINATION-DEPENDENT DNA REPLICATION IN BACTERIOPHAGE T4.ANNUAL REVIEW OF GENETICS. VOL. 32. ISSUE . P. 379 -+ | 42 | 51% | 119 |
| 5 | GRUIDL, ME , CHEN, TC , GARGANO, S , STORLAZZI, A , CASCINO, A , MOSIG, G , (1991) 2 BACTERIOPHAGE-T4 BASE PLATE GENES (25 AND 26) AND THE DNA-REPAIR GENE UVSY BELONG TO SPATIALLY AND TEMPORALLY OVERLAPPING TRANSCRIPTION UNITS.VIROLOGY. VOL. 184. ISSUE 1. P. 359-369 | 35 | 88% | 10 |
| 6 | YONESAKI, T , (1995) RECOMBINATION APPARATUS OF T4 PHAGE.ADVANCES IN BIOPHYSICS, VOL 31, 1995. VOL. 31. ISSUE . P. 3-22 | 36 | 67% | 1 |
| 7 | KUROKI, E , YONESAKI, T , (1999) DNA HELICASE MUTANTS OF BACTERIOPHAGE T4 THAT ARE DEFECTIVE IN DNA RECOMBINATION.MOLECULAR AND GENERAL GENETICS. VOL. 262. ISSUE 3. P. 525 -533 | 25 | 76% | 5 |
| 8 | KREUZER, KN , (2000) RECOMBINATION-DEPENDENT DNA REPLICATION IN PHAGE T4.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 25. ISSUE 4. P. 165 -173 | 24 | 65% | 77 |
| 9 | BIRKENKAMPDEMTRODER, K , GOLZ, S , KEMPER, B , (1997) INHIBITION OF HOLLIDAY STRUCTURE RESOLVING ENDONUCLEASE VII OF BACTERIOPHAGE T4 BY RECOMBINATION ENZYMES UVSX AND UVSY.JOURNAL OF MOLECULAR BIOLOGY. VOL. 267. ISSUE 1. P. 150-162 | 28 | 65% | 7 |
| 10 | FARB, JN , MORRICAL, SW , (2009) FUNCTIONAL COMPLEMENTATION OF UVSX AND UVSY MUTATIONS IN THE MEDIATION OF T4 HOMOLOGOUS RECOMBINATION.NUCLEIC ACIDS RESEARCH. VOL. 37. ISSUE 7. P. 2336 -2345 | 19 | 66% | 2 |
Classes with closest relation at Level 1 |