Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
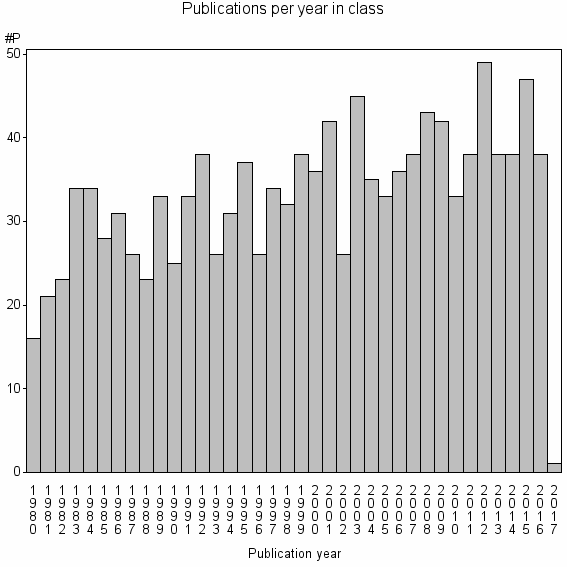
| 8496 | 1247 | 44.0 | 72% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 160 | 2 | WERNER SYNDROME//DNA POLYMERASE//DNA REPAIR | 22798 |
| 8496 | 1 | RECBCD//RECBCD ENZYME//RECOMBINEERING | 1247 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RECBCD | authKW | 511673 | 3% | 65% | 32 |
| 2 | RECBCD ENZYME | authKW | 445186 | 2% | 91% | 20 |
| 3 | RECOMBINEERING | authKW | 419293 | 3% | 41% | 42 |
| 4 | RECF PATHWAY | authKW | 252579 | 1% | 74% | 14 |
| 5 | ADDAB | authKW | 174117 | 1% | 89% | 8 |
| 6 | LAMBDA RED | authKW | 163230 | 1% | 67% | 10 |
| 7 | RECR | authKW | 156704 | 1% | 80% | 8 |
| 8 | RECF | authKW | 141072 | 1% | 52% | 11 |
| 9 | RECFOR | authKW | 88143 | 0% | 60% | 6 |
| 10 | RECJ | authKW | 88143 | 0% | 60% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 6276 | 30% | 0% | 374 |
| 2 | Biochemistry & Molecular Biology | 4387 | 50% | 0% | 618 |
| 3 | Microbiology | 3769 | 24% | 0% | 296 |
| 4 | Biotechnology & Applied Microbiology | 430 | 9% | 0% | 115 |
| 5 | Biochemical Research Methods | 373 | 7% | 0% | 83 |
| 6 | Toxicology | 153 | 4% | 0% | 51 |
| 7 | Biology | 124 | 4% | 0% | 44 |
| 8 | Cell Biology | 110 | 7% | 0% | 86 |
| 9 | Biophysics | 61 | 4% | 0% | 46 |
| 10 | Developmental Biology | 17 | 1% | 0% | 14 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ST PETERSBURG KONSTANTINOV NUCL PHYS | 85692 | 1% | 50% | 7 |
| 2 | MOL CONTROL GENET SECT | 74612 | 1% | 38% | 8 |
| 3 | ES CELL IL | 73456 | 0% | 100% | 3 |
| 4 | BIOINNOVAT ZENTRUM | 58076 | 1% | 22% | 11 |
| 5 | BIOINNOVATIONSZENTRUM | 48971 | 0% | 100% | 2 |
| 6 | SYMBIOSIS FUNCT GENOM UNIT | 48971 | 0% | 100% | 2 |
| 7 | GENE REGULAT CHROMOSOME BIOL | 47953 | 2% | 9% | 21 |
| 8 | AJINOMOTO GENET | 32646 | 0% | 67% | 2 |
| 9 | GENET PL URLGA | 32646 | 0% | 67% | 2 |
| 10 | REGENERAT THER Y D DEN | 32646 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BACTERIOLOGY | 15449 | 12% | 0% | 145 |
| 2 | GENETICS | 9324 | 6% | 1% | 70 |
| 3 | MOLECULAR AND GENERAL GENETICS | 6929 | 3% | 1% | 37 |
| 4 | NUCLEIC ACIDS RESEARCH | 5260 | 7% | 0% | 93 |
| 5 | MOLECULAR & GENERAL GENETICS | 4848 | 2% | 1% | 22 |
| 6 | BMC MOLECULAR BIOLOGY | 3892 | 1% | 1% | 11 |
| 7 | JOURNAL OF MOLECULAR BIOLOGY | 3582 | 5% | 0% | 59 |
| 8 | DNA REPAIR | 3451 | 1% | 1% | 17 |
| 9 | ACS SYNTHETIC BIOLOGY | 2798 | 1% | 1% | 8 |
| 10 | MOLECULAR MICROBIOLOGY | 2331 | 3% | 0% | 34 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RECBCD | 511673 | 3% | 65% | 32 | Search RECBCD | Search RECBCD |
| 2 | RECBCD ENZYME | 445186 | 2% | 91% | 20 | Search RECBCD+ENZYME | Search RECBCD+ENZYME |
| 3 | RECOMBINEERING | 419293 | 3% | 41% | 42 | Search RECOMBINEERING | Search RECOMBINEERING |
| 4 | RECF PATHWAY | 252579 | 1% | 74% | 14 | Search RECF+PATHWAY | Search RECF+PATHWAY |
| 5 | ADDAB | 174117 | 1% | 89% | 8 | Search ADDAB | Search ADDAB |
| 6 | LAMBDA RED | 163230 | 1% | 67% | 10 | Search LAMBDA+RED | Search LAMBDA+RED |
| 7 | RECR | 156704 | 1% | 80% | 8 | Search RECR | Search RECR |
| 8 | RECF | 141072 | 1% | 52% | 11 | Search RECF | Search RECF |
| 9 | RECFOR | 88143 | 0% | 60% | 6 | Search RECFOR | Search RECFOR |
| 10 | RECJ | 88143 | 0% | 60% | 6 | Search RECJ | Search RECJ |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | DILLINGHAM, MS , KOWALCZYKOWSKI, SC , (2008) RECBCD ENZYME AND THE REPAIR OF DOUBLE-STRANDED DNA BREAKS.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 72. ISSUE 4. P. 642-+ | 155 | 62% | 172 |
| 2 | KOWALCZYKOWSKI, SC , DIXON, DA , EGGLESTON, AK , LAUDER, SD , REHRAUER, WM , (1994) BIOCHEMISTRY OF HOMOLOGOUS RECOMBINATION IN ESCHERICHIA-COLI.MICROBIOLOGICAL REVIEWS. VOL. 58. ISSUE 3. P. 401 -465 | 177 | 32% | 928 |
| 3 | KUZMINOV, A , (1999) RECOMBINATIONAL REPAIR OF DNA DAMAGE IN ESCHERICHIA COLI AND BACTERIOPHAGE LAMBDA.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 63. ISSUE 4. P. 751 -+ | 167 | 30% | 584 |
| 4 | MURPHY, KC , (2012) PHAGE RECOMBINASES AND THEIR APPLICATIONS.ADVANCES IN VIRUS RESEARCH, VOL 83: BACTERIOPHAGES, PT B. VOL. 83. ISSUE . P. 367 -414 | 89 | 58% | 20 |
| 5 | COURT, DL , SAWITZKE, JA , THOMASON, LC , (2002) GENETIC ENGINEERING USING HOMOLOGOUS RECOMBINATION.ANNUAL REVIEW OF GENETICS. VOL. 36. ISSUE . P. 361 -388 | 80 | 65% | 243 |
| 6 | AMUNDSEN, SK , SMITH, GR , (2007) CHI HOTSPOT ACTIVITY IN ESCHERICHIA COLI WITHOUT RECBCD EXONUCLEASE ACTIVITY: IMPLICATIONS FOR THE MECHANISM OF RECOMBINATION.GENETICS. VOL. 175. ISSUE 1. P. 41 -54 | 59 | 89% | 7 |
| 7 | REDDY, TR , FEVAT, LMS , MUNSON, SE , STEWART, AF , COWLEY, SM , (2015) LAMBDA RED MEDIATED GAP REPAIR UTILIZES A NOVEL REPLICATIVE INTERMEDIATE IN ESCHERICHIA COLI.PLOS ONE. VOL. 10. ISSUE 3. P. - | 52 | 80% | 1 |
| 8 | SMITH, GR , (2012) HOW RECBCD ENZYME AND CHI PROMOTE DNA BREAK REPAIR AND RECOMBINATION: A MOLECULAR BIOLOGIST'S VIEW.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 76. ISSUE 2. P. 217-228 | 51 | 75% | 29 |
| 9 | AMUNDSEN, SK , SHARP, JW , SMITH, GR , (2016) RECBCD ENZYME "CHI RECOGNITION" MUTANTS RECOGNIZE CHI RECOMBINATION HOTSPOTS IN THE RIGHT DNA CONTEXT.GENETICS. VOL. 204. ISSUE 1. P. 139 -+ | 36 | 100% | 0 |
| 10 | YEELES, JTP , DILLINGHAM, MS , (2010) THE PROCESSING OF DOUBLE-STRANDED DNA BREAKS FOR RECOMBINATIONAL REPAIR BY HELICASE-NUCLEASE COMPLEXES.DNA REPAIR. VOL. 9. ISSUE 3. P. 276 -285 | 67 | 57% | 27 |
Classes with closest relation at Level 1 |