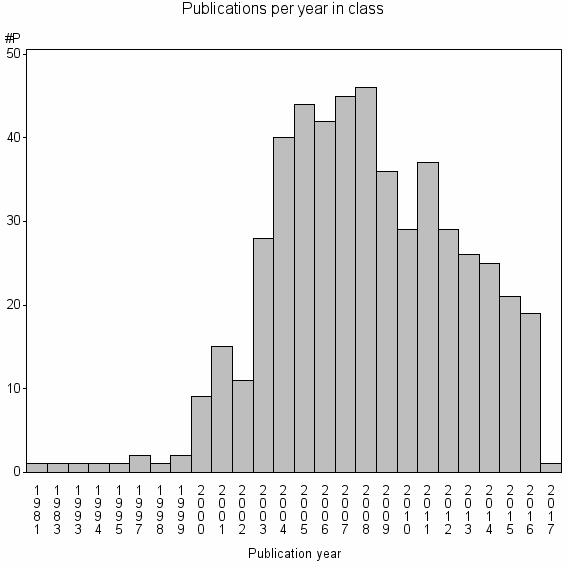
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 19282 | 513 | 46.4 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 19282 | 1 | CONSERVED NON CODING ELEMENTS//IROQUOIS//ENERGY ENVIRONM BIOL COMP | 513 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CONSERVED NON CODING ELEMENTS | authKW | 535686 | 3% | 60% | 15 |
| 2 | IROQUOIS | authKW | 188602 | 3% | 21% | 15 |
| 3 | ENERGY ENVIRONM BIOL COMP | address | 178566 | 1% | 100% | 3 |
| 4 | SARS MARINE MOL BIOL | address | 145765 | 3% | 19% | 13 |
| 5 | ULTRACONSERVED ELEMENTS | authKW | 137738 | 2% | 26% | 9 |
| 6 | GENOMIC REGULATORY BLOCKS | authKW | 119044 | 0% | 100% | 2 |
| 7 | ULTRACONSERVED ELEMENTS UCES | authKW | 119044 | 0% | 100% | 2 |
| 8 | ZEBRAFISH TRANSGENESIS | authKW | 119044 | 0% | 100% | 2 |
| 9 | CNE | authKW | 105812 | 1% | 44% | 4 |
| 10 | ENHANCER EVOLUTION | authKW | 86572 | 1% | 36% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 10221 | 58% | 0% | 298 |
| 2 | Biotechnology & Applied Microbiology | 3498 | 35% | 0% | 181 |
| 3 | Developmental Biology | 1600 | 12% | 0% | 59 |
| 4 | Evolutionary Biology | 1276 | 11% | 0% | 55 |
| 5 | Biochemistry & Molecular Biology | 1079 | 40% | 0% | 203 |
| 6 | Mathematical & Computational Biology | 867 | 8% | 0% | 40 |
| 7 | Biochemical Research Methods | 291 | 9% | 0% | 45 |
| 8 | Biology | 66 | 4% | 0% | 20 |
| 9 | Cell Biology | 60 | 8% | 0% | 39 |
| 10 | Anatomy & Morphology | 38 | 1% | 0% | 7 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ENERGY ENVIRONM BIOL COMP | 178566 | 1% | 100% | 3 |
| 2 | SARS MARINE MOL BIOL | 145765 | 3% | 19% | 13 |
| 3 | PROGRAM COMPARAT EVOLUTIONARY GENOM | 73252 | 1% | 31% | 4 |
| 4 | BIOL DESAROLLO | 59522 | 0% | 100% | 1 |
| 5 | BIOL MOL SEVERO OATOA | 59522 | 0% | 100% | 1 |
| 6 | CANC AGINGINSERMU1081 | 59522 | 0% | 100% | 1 |
| 7 | CSIC UPO JA | 59522 | 0% | 100% | 1 |
| 8 | DEV BIOLCHUOU KU | 59522 | 0% | 100% | 1 |
| 9 | DEV NEUROBIOL GENET | 59522 | 0% | 100% | 1 |
| 10 | DEV TRANSCRIPT REGULAT | 59522 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOTECHFORUM ( BTF ) | 34986 | 8% | 2% | 39 |
| 2 | GENOME RESEARCH | 11058 | 3% | 1% | 16 |
| 3 | GENOME BIOLOGY AND EVOLUTION | 9905 | 3% | 1% | 15 |
| 4 | MOLECULAR BIOLOGY AND EVOLUTION | 3830 | 4% | 0% | 19 |
| 5 | GENOME BIOLOGY | 3353 | 3% | 0% | 13 |
| 6 | TRENDS IN GENETICS | 3137 | 2% | 0% | 12 |
| 7 | ANNUAL REVIEW OF GENOMICS AND HUMAN GENETICS | 2959 | 1% | 1% | 4 |
| 8 | GENOMICS | 2854 | 4% | 0% | 20 |
| 9 | BMC GENOMICS | 2462 | 4% | 0% | 20 |
| 10 | BIOINFORMATICS | 1990 | 4% | 0% | 19 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CONSERVED NON CODING ELEMENTS | 535686 | 3% | 60% | 15 | Search CONSERVED+NON+CODING+ELEMENTS | Search CONSERVED+NON+CODING+ELEMENTS |
| 2 | IROQUOIS | 188602 | 3% | 21% | 15 | Search IROQUOIS | Search IROQUOIS |
| 3 | ULTRACONSERVED ELEMENTS | 137738 | 2% | 26% | 9 | Search ULTRACONSERVED+ELEMENTS | Search ULTRACONSERVED+ELEMENTS |
| 4 | GENOMIC REGULATORY BLOCKS | 119044 | 0% | 100% | 2 | Search GENOMIC+REGULATORY+BLOCKS | Search GENOMIC+REGULATORY+BLOCKS |
| 5 | ULTRACONSERVED ELEMENTS UCES | 119044 | 0% | 100% | 2 | Search ULTRACONSERVED+ELEMENTS+UCES | Search ULTRACONSERVED+ELEMENTS+UCES |
| 6 | ZEBRAFISH TRANSGENESIS | 119044 | 0% | 100% | 2 | Search ZEBRAFISH+TRANSGENESIS | Search ZEBRAFISH+TRANSGENESIS |
| 7 | CNE | 105812 | 1% | 44% | 4 | Search CNE | Search CNE |
| 8 | ENHANCER EVOLUTION | 86572 | 1% | 36% | 4 | Search ENHANCER+EVOLUTION | Search ENHANCER+EVOLUTION |
| 9 | IRX GENES | 79361 | 0% | 67% | 2 | Search IRX+GENES | Search IRX+GENES |
| 10 | 4 FOLD DEGENERATE SITES | 59522 | 0% | 100% | 1 | Search 4+FOLD+DEGENERATE+SITES | Search 4+FOLD+DEGENERATE+SITES |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | HARMSTON, N , BARESIC, A , LENHARD, B , (2013) THE MYSTERY OF EXTREME NON-CODING CONSERVATION.PHILOSOPHICAL TRANSACTIONS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES. VOL. 368. ISSUE 1632. P. - | 66 | 57% | 14 |
| 2 | SAKURABA, Y , KIMURA, T , MASUYA, H , NOGUCHI, H , SEZUTSU, H , TAKAHASI, KR , TOYODA, A , FUKUMURA, R , MURATA, T , SAKAKI, Y , ET AL (2008) IDENTIFICATION AND CHARACTERIZATION OF NEW LONG CONSERVED NONCODING SEQUENCES IN VERTEBRATES.MAMMALIAN GENOME. VOL. 19. ISSUE 10-12. P. 703-712 | 34 | 74% | 18 |
| 3 | POLYCHRONOPOULOS, D , SELLIS, D , ALMIRANTIS, Y , (2014) CONSERVED NONCODING ELEMENTS FOLLOW POWER-LAW-LIKE DISTRIBUTIONS IN SEVERAL GENOMES AS A RESULT OF GENOME DYNAMICS.PLOS ONE. VOL. 9. ISSUE 5. P. - | 39 | 51% | 1 |
| 4 | SIEPEL, A , BEJERANO, G , PEDERSEN, JS , HINRICHS, AS , HOU, MM , ROSENBLOOM, K , CLAWSON, H , SPIETH, J , HILLIER, LW , RICHARDS, S , ET AL (2005) EVOLUTIONARILY CONSERVED ELEMENTS IN VERTEBRATE, INSECT, WORM, AND YEAST GENOMES.GENOME RESEARCH. VOL. 15. ISSUE 8. P. 1034 -1050 | 25 | 32% | 1514 |
| 5 | WOOLFE, A , ELGAR, G , (2008) ORGANIZATION OF CONSERVED ELEMENTS NEAR KEY DEVELOPMENTAL REGULATORS IN VERTEBRATE GENOMES.LONG-RANGE CONTROL OF GENE EXPRESSION. VOL. 61. ISSUE . P. 307-338 | 38 | 45% | 22 |
| 6 | VENKATESH, B , MAESO, I , RAVI, V , IRIMIA, M , TENA, JJ , GONZALEZ-PEREZ, E , TRAN, D , CAMPUZANO, S , GOMEZ-SKARMETA, JL , GARCIA-FERNANDEZ, J , (2012) AN ANCIENT GENOMIC REGULATORY BLOCK CONSERVED ACROSS BILATERIANS AND ITS DISMANTLING IN TETRAPODS BY RETROGENE REPLACEMENT.GENOME RESEARCH. VOL. 22. ISSUE 4. P. 642 -655 | 34 | 46% | 6 |
| 7 | POLLARD, KS , HUBISZ, MJ , ROSENBLOOM, KR , SIEPEL, A , (2010) DETECTION OF NONNEUTRAL SUBSTITUTION RATES ON MAMMALIAN PHYLOGENIES.GENOME RESEARCH. VOL. 20. ISSUE 1. P. 110-121 | 21 | 38% | 415 |
| 8 | DIMITRIEVA, S , BUCHER, P , (2013) UCNEBASE-A DATABASE OF ULTRACONSERVED NON-CODING ELEMENTS AND GENOMIC REGULATORY BLOCKS.NUCLEIC ACIDS RESEARCH. VOL. 41. ISSUE D1. P. D101 -D109 | 17 | 74% | 15 |
| 9 | ROYO, JL , HIDALGO, C , RONCERO, Y , SEDA, MA , AKALIN, A , LENHARD, B , CASARES, F , GOMEZ-SKARMETA, JL , (2011) DISSECTING THE TRANSCRIPTIONAL REGULATORY PROPERTIES OF HUMAN CHROMOSOME 16 HIGHLY CONSERVED NON-CODING REGIONS.PLOS ONE. VOL. 6. ISSUE 9. P. - | 24 | 57% | 5 |
| 10 | RITTER, DI , LI, QA , KOSTKA, D , POLLARD, KS , GUO, S , CHUANG, JH , (2010) THE IMPORTANCE OF BEING CIS: EVOLUTION OF ORTHOLOGOUS FISH AND MAMMALIAN ENHANCER ACTIVITY.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 27. ISSUE 10. P. 2322 -2332 | 23 | 58% | 21 |
Classes with closest relation at Level 1 |