Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
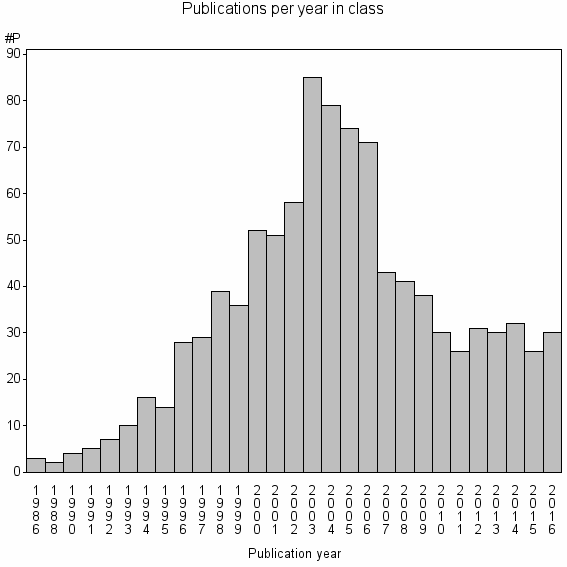
| 11342 | 990 | 30.8 | 76% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 11342 | 1 | GENE PREDICTION//GENE FINDING//GENE RECOGNITION | 990 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENE PREDICTION | authKW | 468962 | 4% | 36% | 42 |
| 2 | GENE FINDING | authKW | 192148 | 2% | 35% | 18 |
| 3 | GENE RECOGNITION | authKW | 166541 | 1% | 60% | 9 |
| 4 | SPLICE SITE PREDICTION | authKW | 140986 | 1% | 57% | 8 |
| 5 | SPLICE SITE | authKW | 113915 | 2% | 17% | 22 |
| 6 | RE ANNOTATION | authKW | 96378 | 1% | 63% | 5 |
| 7 | P1 GENOMIC LIBRARY | authKW | 92527 | 0% | 100% | 3 |
| 8 | SHANDONG PROV BIOPHYS FUNCT MACROMOL | address | 92527 | 0% | 100% | 3 |
| 9 | EXON PREDICTION | authKW | 85403 | 1% | 46% | 6 |
| 10 | SPLICED ALIGNMENT | authKW | 69394 | 0% | 75% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 26593 | 30% | 0% | 298 |
| 2 | Biotechnology & Applied Microbiology | 9923 | 42% | 0% | 420 |
| 3 | Biochemical Research Methods | 7036 | 28% | 0% | 281 |
| 4 | Genetics & Heredity | 3197 | 24% | 0% | 241 |
| 5 | Biochemistry & Molecular Biology | 1461 | 34% | 0% | 337 |
| 6 | Computer Science, Interdisciplinary Applications | 792 | 8% | 0% | 81 |
| 7 | Biology | 215 | 5% | 0% | 48 |
| 8 | Computer Science, Artificial Intelligence | 142 | 4% | 0% | 37 |
| 9 | Statistics & Probability | 95 | 3% | 0% | 29 |
| 10 | Computer Science, Information Systems | 53 | 3% | 0% | 25 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SHANDONG PROV BIOPHYS FUNCT MACROMOL | 92527 | 0% | 100% | 3 |
| 2 | 659 | 61685 | 0% | 100% | 2 |
| 3 | NUTR DOENCAS METAB | 61685 | 0% | 100% | 2 |
| 4 | INRA FRANCE | 46260 | 0% | 50% | 3 |
| 5 | ASSOCIE INRA FRANCE | 41122 | 0% | 67% | 2 |
| 6 | AG PRAKT INFORMAT | 30842 | 0% | 100% | 1 |
| 7 | ANAL EXP SAO GENICA | 30842 | 0% | 100% | 1 |
| 8 | ATTAT | 30842 | 0% | 100% | 1 |
| 9 | BIO COMP IL | 30842 | 0% | 100% | 1 |
| 10 | BIOINFORMAT ERA7 | 30842 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 45967 | 13% | 1% | 126 |
| 2 | BIOTECHFORUM ( BTF ) | 41466 | 6% | 2% | 59 |
| 3 | BMC BIOINFORMATICS | 20077 | 7% | 1% | 70 |
| 4 | COMPUTER APPLICATIONS IN THE BIOSCIENCES | 12212 | 2% | 2% | 19 |
| 5 | JOURNAL OF COMPUTATIONAL BIOLOGY | 11363 | 2% | 2% | 24 |
| 6 | NUCLEIC ACIDS RESEARCH | 9724 | 11% | 0% | 112 |
| 7 | GENOME BIOLOGY | 6426 | 3% | 1% | 25 |
| 8 | CURRENT BIOINFORMATICS | 4450 | 1% | 2% | 8 |
| 9 | DNA RESEARCH | 3910 | 1% | 1% | 9 |
| 10 | COMPUTERS & CHEMISTRY | 3794 | 1% | 1% | 11 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENE PREDICTION | 468962 | 4% | 36% | 42 | Search GENE+PREDICTION | Search GENE+PREDICTION |
| 2 | GENE FINDING | 192148 | 2% | 35% | 18 | Search GENE+FINDING | Search GENE+FINDING |
| 3 | GENE RECOGNITION | 166541 | 1% | 60% | 9 | Search GENE+RECOGNITION | Search GENE+RECOGNITION |
| 4 | SPLICE SITE PREDICTION | 140986 | 1% | 57% | 8 | Search SPLICE+SITE+PREDICTION | Search SPLICE+SITE+PREDICTION |
| 5 | SPLICE SITE | 113915 | 2% | 17% | 22 | Search SPLICE+SITE | Search SPLICE+SITE |
| 6 | RE ANNOTATION | 96378 | 1% | 63% | 5 | Search RE+ANNOTATION | Search RE+ANNOTATION |
| 7 | P1 GENOMIC LIBRARY | 92527 | 0% | 100% | 3 | Search P1+GENOMIC+LIBRARY | Search P1+GENOMIC+LIBRARY |
| 8 | EXON PREDICTION | 85403 | 1% | 46% | 6 | Search EXON+PREDICTION | Search EXON+PREDICTION |
| 9 | SPLICED ALIGNMENT | 69394 | 0% | 75% | 3 | Search SPLICED+ALIGNMENT | Search SPLICED+ALIGNMENT |
| 10 | ARABIDOPSIS THALIANA CHROMOSOME 3 | 61685 | 0% | 100% | 2 | Search ARABIDOPSIS+THALIANA+CHROMOSOME+3 | Search ARABIDOPSIS+THALIANA+CHROMOSOME+3 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MATHE, C , SAGOT, MF , SCHIEX, T , ROUZE, P , (2002) CURRENT METHODS OF GENE PREDICTION, THEIR STRENGTHS AND WEAKNESSES.NUCLEIC ACIDS RESEARCH. VOL. 30. ISSUE 19. P. 4103 -4117 | 82 | 62% | 215 |
| 2 | BRENT, MR , (2008) STEADY PROGRESS AND RECENT BREAKTHROUGHS IN THE ACCURACY OF AUTOMATED GENOME ANNOTATION.NATURE REVIEWS GENETICS. VOL. 9. ISSUE 1. P. 62 -73 | 35 | 69% | 77 |
| 3 | HAAS, BJ , SALZBERG, SL , ZHU, W , PERTEA, M , ALLEN, JE , ORVIS, J , WHITE, O , BUELL, CR , WORTMAN, JR , (2008) AUTOMATED EUKARYOTIC GENE STRUCTURE ANNOTATION USING EVIDENCEMODELER AND THE PROGRAM TO ASSEMBLE SPLICED ALIGNMENTS.GENOME BIOLOGY. VOL. 9. ISSUE 1. P. - | 27 | 77% | 160 |
| 4 | LIU, Q , MACKEY, AJ , ROOS, DS , PEREIRA, FCN , (2008) EVIGAN: A HIDDEN VARIABLE MODEL FOR INTEGRATING GENE EVIDENCE FOR EUKARYOTIC GENE PREDICTION.BIOINFORMATICS. VOL. 24. ISSUE 5. P. 597 -605 | 28 | 93% | 17 |
| 5 | GOEL, N , SINGH, S , ASERI, TC , (2013) A COMPARATIVE ANALYSIS OF SOFT COMPUTING TECHNIQUES FOR GENE PREDICTION.ANALYTICAL BIOCHEMISTRY. VOL. 438. ISSUE 1. P. 14-21 | 28 | 80% | 4 |
| 6 | MAJI, S , GARG, D , (2013) PROGRESS IN GENE PREDICTION: PRINCIPLES AND CHALLENGES.CURRENT BIOINFORMATICS. VOL. 8. ISSUE 2. P. 226 -243 | 41 | 55% | 3 |
| 7 | LOMSADZE, A , TER-HOVHANNISYAN, V , CHERNOFF, YO , BORODOVSKY, M , (2005) GENE IDENTIFICATION IN NOVEL EUKARYOTIC GENOMES BY SELF-TRAINING ALGORITHM.NUCLEIC ACIDS RESEARCH. VOL. 33. ISSUE 20. P. 6494-6506 | 28 | 78% | 173 |
| 8 | GOODSWEN, SJ , KENNEDY, PJ , ELLIS, JT , (2012) EVALUATING HIGH-THROUGHPUT AB INITIO GENE FINDERS TO DISCOVER PROTEINS ENCODED IN EUKARYOTIC PATHOGEN GENOMES MISSED BY LABORATORY TECHNIQUES.PLOS ONE. VOL. 7. ISSUE 11. P. - | 33 | 66% | 6 |
| 9 | CHURBANOV, A , (2006) CONTEMPORARY PROGRESS IN GENE STRUCTURE PREDICTION.CURRENT GENOMICS. VOL. 7. ISSUE 5. P. 283-292 | 38 | 67% | 0 |
| 10 | FOISSAC, S , GOUZY, J , ROMBAUTS, S , MATHE, C , AMSELEM, J , STERCK, L , VAN DE PEER, Y , ROUZE, P , SCHIEX, T , (2008) GENOME ANNOTATION IN PLANTS AND FUNGI: EUGENE AS A MODEL PLATFORM.CURRENT BIOINFORMATICS. VOL. 3. ISSUE 2. P. 87 -97 | 30 | 75% | 15 |
Classes with closest relation at Level 1 |