Methods for the analysis and characterization of brain morphology from MRI images
Description
Brain magnetic resonance imaging (MRI) is one of the most commonly used methods for investigating brain morphology, since it allows to produce detailed 3D images of its soft-tissue structures. In this project, we are working on the development of methods for the analysis and characterisation of brain morphology from both structural and diffusion brain MRI images.
In our first work, we investigated the differences between the brains of domestic and wild rabbits from high resolution MRI images. In particular, structural T1-weighted scans were processed by carrying out atlas-based segmentation and voxel-based morphometry, while diffusion MRI images were analysed by performing tract-based spatial statistics. The results identified that domestic rabbits have an altered brain architecture, which is consistent with reduced emotional processing (including fear detection, learning, expression, and control), as well as compromised information processing.
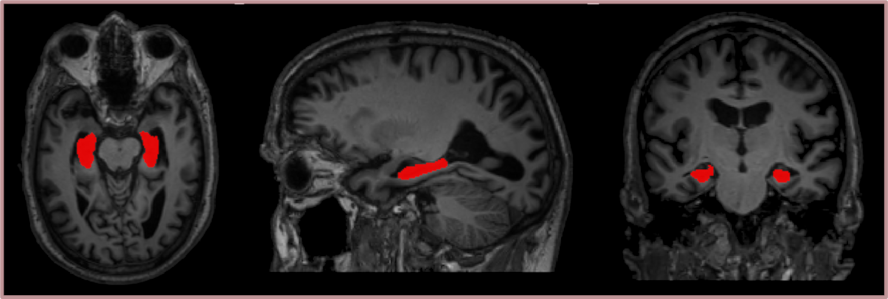
Our following works have instead been focusing on the development of image processing tools that can be employed to identify and analyse neurodegenerative patterns in the brains of dementia patients, with a special focus on Alzheimer’s patients. Our methods are being tested in several large databases, available at the Geriatrics unit of Karolinska university hospital Huddinge and containing image data, from patients with several types of dementia as well as from normal controls. In particular, we developed an automatic hippocampal segmentation tool that integrates a statistical shape model of the hippocampus into a traditional deep learning-based segmentation pipeline. This method achieves a high accuracy not only on healthy controls, but also on Alzheimer’s patients where automatic segmentation is usually more challenging due to their hippocampal atrophy. Moreover, we are currently working on the creation of a deep learning-based tool for brain segmentation quality control.
Short presentation video about our on-going work (presented at the Organization for Human Brain Mapping Meeting 2020):
Publications
- I. Brusini et al., "Shape Information Improves the Cross-Cohort Performance of Deep Learning-Based Segmentation of the Hippocampus," Frontiers in Neuroscience, vol. 14, 2020.
- I. Brusini et al., "Changes in brain architecture are consistent with altered fear processing in domestic rabbits," Proceedings of the National Academy of Sciences of the United States of America, vol. 115, no. 28, pp. 7380-7385, 2018.
Collaborations
- Geriatrics, KI
- Geriatics, Karolinska university hospital Huddinge
Thesis projects
Irene Brusini, doctoral dissertation project, ongoing.
