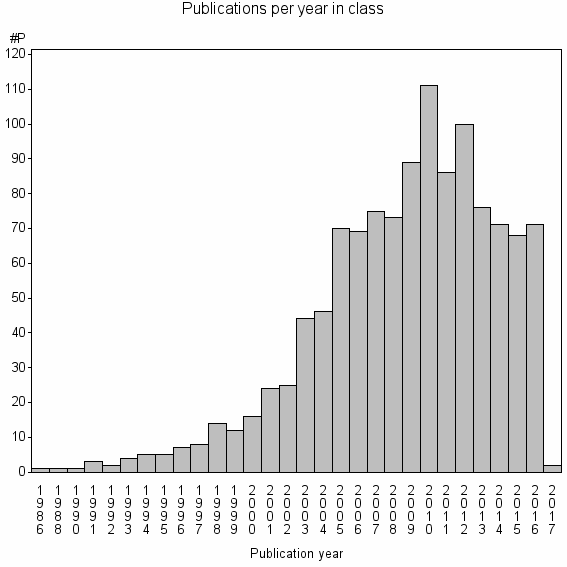
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 9196 | 1179 | 44.5 | 85% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 9196 | 1 | PROTEIN FUNCTION PREDICTION//COMPLEX SCI SOFTWARE//PHYLOGENETIC MOTIFS | 1179 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN FUNCTION PREDICTION | authKW | 268983 | 3% | 27% | 38 |
| 2 | COMPLEX SCI SOFTWARE | address | 190698 | 1% | 82% | 9 |
| 3 | PHYLOGENETIC MOTIFS | authKW | 155387 | 1% | 100% | 6 |
| 4 | THEMATICS | authKW | 139841 | 1% | 60% | 9 |
| 5 | STRUCTURE BASED FUNCTION PREDICTION | authKW | 133187 | 1% | 86% | 6 |
| 6 | FUNCTIONAL SITE PREDICTION | authKW | 118383 | 1% | 57% | 8 |
| 7 | PROTEIN FUNCTION | authKW | 118319 | 3% | 11% | 40 |
| 8 | BINDING SITE COMPARISON | authKW | 107906 | 0% | 83% | 5 |
| 9 | STRUCTURAL GENOMICS | authKW | 97034 | 4% | 8% | 45 |
| 10 | STUDY SYST BIOL | address | 96418 | 2% | 21% | 18 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 27003 | 28% | 0% | 328 |
| 2 | Biochemical Research Methods | 8660 | 29% | 0% | 340 |
| 3 | Biochemistry & Molecular Biology | 4272 | 50% | 0% | 592 |
| 4 | Biotechnology & Applied Microbiology | 3444 | 24% | 0% | 279 |
| 5 | Computer Science, Interdisciplinary Applications | 2234 | 12% | 0% | 144 |
| 6 | Biophysics | 1960 | 16% | 0% | 183 |
| 7 | Chemistry, Medicinal | 682 | 8% | 0% | 92 |
| 8 | Computer Science, Information Systems | 474 | 6% | 0% | 68 |
| 9 | Statistics & Probability | 79 | 3% | 0% | 30 |
| 10 | Chemistry, Multidisciplinary | 67 | 8% | 0% | 95 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | COMPLEX SCI SOFTWARE | 190698 | 1% | 82% | 9 |
| 2 | STUDY SYST BIOL | 96418 | 2% | 21% | 18 |
| 3 | GLOBAL COMPUTAT CHEM | 84751 | 1% | 55% | 6 |
| 4 | MOL BIOINFORMAT | 64719 | 1% | 17% | 15 |
| 5 | BIOMED SCI COMPLEX EASTCHEM CHEM | 51796 | 0% | 100% | 2 |
| 6 | MPI COMPUTAT BIOL | 51796 | 0% | 100% | 2 |
| 7 | CIBR COMPUTAT INTEGRAT BIOMED | 34529 | 0% | 67% | 2 |
| 8 | OPEN SOURCE DRUG DISCOVERY OSDD CONSORTIUM | 34529 | 0% | 67% | 2 |
| 9 | ADV IT DEV | 25898 | 0% | 100% | 1 |
| 10 | ARC STRUCT FUNCT MICROBIAL GENET | 25898 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 34962 | 10% | 1% | 120 |
| 2 | BMC BIOINFORMATICS | 33076 | 8% | 1% | 98 |
| 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 32094 | 8% | 1% | 89 |
| 4 | JOURNAL OF CHEMICAL INFORMATION AND MODELING | 26080 | 5% | 2% | 56 |
| 5 | BMC STRUCTURAL BIOLOGY | 18624 | 2% | 4% | 19 |
| 6 | JOURNAL OF MOLECULAR BIOLOGY | 8141 | 7% | 0% | 86 |
| 7 | NUCLEIC ACIDS RESEARCH | 7552 | 9% | 0% | 108 |
| 8 | PLOS COMPUTATIONAL BIOLOGY | 5336 | 3% | 1% | 31 |
| 9 | PROTEIN SCIENCE | 4448 | 3% | 1% | 33 |
| 10 | IEEE-ACM TRANSACTIONS ON COMPUTATIONAL BIOLOGY AND BIOINFORMATICS | 3848 | 1% | 1% | 13 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN FUNCTION PREDICTION | 268983 | 3% | 27% | 38 | Search PROTEIN+FUNCTION+PREDICTION | Search PROTEIN+FUNCTION+PREDICTION |
| 2 | PHYLOGENETIC MOTIFS | 155387 | 1% | 100% | 6 | Search PHYLOGENETIC+MOTIFS | Search PHYLOGENETIC+MOTIFS |
| 3 | THEMATICS | 139841 | 1% | 60% | 9 | Search THEMATICS | Search THEMATICS |
| 4 | STRUCTURE BASED FUNCTION PREDICTION | 133187 | 1% | 86% | 6 | Search STRUCTURE+BASED+FUNCTION+PREDICTION | Search STRUCTURE+BASED+FUNCTION+PREDICTION |
| 5 | FUNCTIONAL SITE PREDICTION | 118383 | 1% | 57% | 8 | Search FUNCTIONAL+SITE+PREDICTION | Search FUNCTIONAL+SITE+PREDICTION |
| 6 | PROTEIN FUNCTION | 118319 | 3% | 11% | 40 | Search PROTEIN+FUNCTION | Search PROTEIN+FUNCTION |
| 7 | BINDING SITE COMPARISON | 107906 | 0% | 83% | 5 | Search BINDING+SITE+COMPARISON | Search BINDING+SITE+COMPARISON |
| 8 | STRUCTURAL GENOMICS | 97034 | 4% | 8% | 45 | Search STRUCTURAL+GENOMICS | Search STRUCTURAL+GENOMICS |
| 9 | PROTEIN SURFACE SHAPE | 92489 | 0% | 71% | 5 | Search PROTEIN+SURFACE+SHAPE | Search PROTEIN+SURFACE+SHAPE |
| 10 | FUNCTIONAL RESIDUES | 92072 | 1% | 44% | 8 | Search FUNCTIONAL+RESIDUES | Search FUNCTIONAL+RESIDUES |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | JALENCAS, X , MESTRES, J , (2013) IDENTIFICATION OF SIMILAR BINDING SITES TO DETECT DISTANT POLYPHARMACOLOGY.MOLECULAR INFORMATICS. VOL. 32. ISSUE 11-12. P. 976-990 | 93 | 55% | 8 |
| 2 | NISIUS, B , SHA, F , GOHLKE, H , (2012) STRUCTURE-BASED COMPUTATIONAL ANALYSIS OF PROTEIN BINDING SITES FOR FUNCTION AND DRUGGABILITY PREDICTION.JOURNAL OF BIOTECHNOLOGY. VOL. 159. ISSUE 3. P. 123-134 | 55 | 71% | 40 |
| 3 | VOLKAMER, A , RAREY, M , (2014) EXPLOITING STRUCTURAL INFORMATION FOR DRUG- TARGET ASSESSMENT.FUTURE MEDICINAL CHEMISTRY. VOL. 6. ISSUE 3. P. 319-331 | 58 | 68% | 5 |
| 4 | CHAKRABORTY, A , CHAKRABARTI, S , (2015) A SURVEY ON PREDICTION OF SPECIFICITY-DETERMINING SITES IN PROTEINS.BRIEFINGS IN BIOINFORMATICS. VOL. 16. ISSUE 1. P. 71 -88 | 48 | 75% | 6 |
| 5 | CAPRA, JA , LASKOWSKI, RA , THORNTON, JM , SINGH, M , FUNKHOUSER, TA , (2009) PREDICTING PROTEIN LIGAND BINDING SITES BY COMBINING EVOLUTIONARY SEQUENCE CONSERVATION AND 3D STRUCTURE.PLOS COMPUTATIONAL BIOLOGY. VOL. 5. ISSUE 12. P. - | 47 | 73% | 131 |
| 6 | XIN, FX , RADIVOJAC, P , (2011) COMPUTATIONAL METHODS FOR IDENTIFICATION OF FUNCTIONAL RESIDUES IN PROTEIN STRUCTURES.CURRENT PROTEIN & PEPTIDE SCIENCE. VOL. 12. ISSUE 6. P. 456-469 | 69 | 58% | 8 |
| 7 | PEROT, S , SPERANDIO, O , MITEVA, MA , CAMPROUX, AC , VILLOUTREIX, BO , (2010) DRUGGABLE POCKETS AND BINDING SITE CENTRIC CHEMICAL SPACE: A PARADIGM SHIFT IN DRUG DISCOVERY.DRUG DISCOVERY TODAY. VOL. 15. ISSUE 15-16. P. 656 -667 | 58 | 51% | 111 |
| 8 | BRAY, T , CHAN, P , BOUGOUFFA, S , GREAVES, R , DOIG, AJ , WARWICKER, J , (2009) SITESIDENTIFY: A PROTEIN FUNCTIONAL SITE PREDICTION TOOL.BMC BIOINFORMATICS. VOL. 10. ISSUE . P. - | 47 | 81% | 13 |
| 9 | PRYMULA, K , JADCZYK, T , ROTERMAN, I , (2011) CATALYTIC RESIDUES IN HYDROLASES: ANALYSIS OF METHODS DESIGNED FOR LIGAND-BINDING SITE PREDICTION.JOURNAL OF COMPUTER-AIDED MOLECULAR DESIGN. VOL. 25. ISSUE 2. P. 117-133 | 55 | 59% | 6 |
| 10 | ZHANG, ZD , TANG, YR , SHENG, ZY , ZHAO, DB , (2009) AN OVERVIEW OF THE DE NOVO PREDICTION OF ENZYME CATALYTIC RESIDUES.CURRENT BIOINFORMATICS. VOL. 4. ISSUE 3. P. 197-206 | 49 | 66% | 2 |
Classes with closest relation at Level 1 |