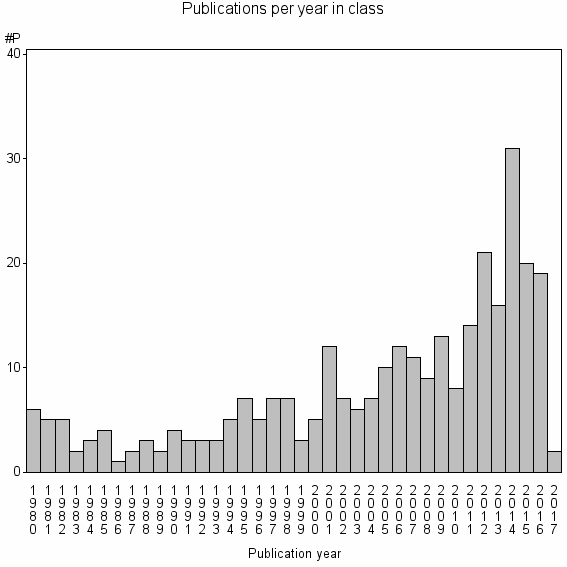
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 24744 | 303 | 34.6 | 72% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 278 | 3 | CORYNEBACTERIUM GLUTAMICUM//ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY//BIOCHEMISTRY & MOLECULAR BIOLOGY | 43163 |
| 2622 | 2 | TRIOSEPHOSPHATE ISOMERASE//HISTIDINE BIOSYNTHESIS//KETOSTEROID ISOMERASE | 3550 |
| 24744 | 1 | MANDELATE RACEMASE//MANDELATE RACEMASE EC 5122//ENOLASE SUPERFAMILY | 303 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MANDELATE RACEMASE | authKW | 806202 | 4% | 67% | 12 |
| 2 | MANDELATE RACEMASE EC 5122 | authKW | 403105 | 1% | 100% | 4 |
| 3 | ENOLASE SUPERFAMILY | authKW | 403101 | 2% | 67% | 6 |
| 4 | ENZYMATIC RACEMIZATION | authKW | 302329 | 1% | 100% | 3 |
| 5 | N ACYLAMINO ACID RACEMASE | authKW | 279930 | 2% | 56% | 5 |
| 6 | L RHAMNOSE CATABOLISM | authKW | 226745 | 1% | 75% | 3 |
| 7 | N SUCCINYLAMINO ACID RACEMASE | authKW | 226745 | 1% | 75% | 3 |
| 8 | PSEUDOMONAS PUTIDA ATCC 12633 | authKW | 226745 | 1% | 75% | 3 |
| 9 | L RHAMNONATE | authKW | 201553 | 1% | 100% | 2 |
| 10 | MESO GALACTARIC ACID | authKW | 201553 | 1% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 1407 | 56% | 0% | 170 |
| 2 | Biotechnology & Applied Microbiology | 866 | 23% | 0% | 71 |
| 3 | Biochemical Research Methods | 123 | 8% | 0% | 23 |
| 4 | Mathematical & Computational Biology | 100 | 4% | 0% | 11 |
| 5 | Microbiology | 91 | 9% | 0% | 26 |
| 6 | Biophysics | 80 | 7% | 0% | 21 |
| 7 | Mycology | 41 | 2% | 0% | 5 |
| 8 | Chemistry, Multidisciplinary | 14 | 8% | 0% | 23 |
| 9 | Chemistry, Organic | 5 | 4% | 0% | 11 |
| 10 | Food Science & Technology | 5 | 3% | 0% | 8 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NEW YORK SGX STRUCT GENOM | 151161 | 1% | 50% | 3 |
| 2 | ADV MAT NUCL TECHNOL PL BIO NANOTECHNOL | 100776 | 0% | 100% | 1 |
| 3 | ADV SYNTH CATALYIS | 100776 | 0% | 100% | 1 |
| 4 | BIOL BIOPHYS PROGRAM | 100776 | 0% | 100% | 1 |
| 5 | CONSOLIDATED UNIT UIC 154 | 100776 | 0% | 100% | 1 |
| 6 | INT CRIT RAW MAT ICCRAM | 100776 | 0% | 100% | 1 |
| 7 | QUANTITATIVE BIOSCI | 100776 | 0% | 100% | 1 |
| 8 | ABORATORY STRUCT BIOINFORMAT PROT DATA BA | 80619 | 1% | 40% | 2 |
| 9 | LILLY BIOTECHNOL | 57580 | 1% | 14% | 4 |
| 10 | DEV PLICAT TECHNOL | 50387 | 0% | 50% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOCHEMISTRY | 15442 | 30% | 0% | 90 |
| 2 | HUMAN GENOME NEWS | 3052 | 0% | 3% | 1 |
| 3 | ZEITSCHRIFT FUR ALLGEMEINE MIKROBIOLOGIE | 2426 | 1% | 1% | 3 |
| 4 | JOURNAL OF THE AGRICULTURAL CHEMICAL SOCIETY OF JAPAN | 2113 | 1% | 1% | 3 |
| 5 | AMB EXPRESS | 1738 | 1% | 1% | 3 |
| 6 | FUNGAL GENETICS AND BIOLOGY | 1259 | 2% | 0% | 5 |
| 7 | JOURNAL OF MICRONUTRIENT ANALYSIS | 728 | 0% | 1% | 1 |
| 8 | APPLIED MICROBIOLOGY AND BIOTECHNOLOGY | 713 | 3% | 0% | 10 |
| 9 | BIOTECHNOLOGY LETTERS | 678 | 3% | 0% | 8 |
| 10 | MICROBIAL BIOTECHNOLOGY | 678 | 1% | 0% | 2 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MANDELATE RACEMASE | 806202 | 4% | 67% | 12 | Search MANDELATE+RACEMASE | Search MANDELATE+RACEMASE |
| 2 | MANDELATE RACEMASE EC 5122 | 403105 | 1% | 100% | 4 | Search MANDELATE+RACEMASE+EC+5122 | Search MANDELATE+RACEMASE+EC+5122 |
| 3 | ENOLASE SUPERFAMILY | 403101 | 2% | 67% | 6 | Search ENOLASE+SUPERFAMILY | Search ENOLASE+SUPERFAMILY |
| 4 | ENZYMATIC RACEMIZATION | 302329 | 1% | 100% | 3 | Search ENZYMATIC+RACEMIZATION | Search ENZYMATIC+RACEMIZATION |
| 5 | N ACYLAMINO ACID RACEMASE | 279930 | 2% | 56% | 5 | Search N+ACYLAMINO+ACID+RACEMASE | Search N+ACYLAMINO+ACID+RACEMASE |
| 6 | L RHAMNOSE CATABOLISM | 226745 | 1% | 75% | 3 | Search L+RHAMNOSE+CATABOLISM | Search L+RHAMNOSE+CATABOLISM |
| 7 | N SUCCINYLAMINO ACID RACEMASE | 226745 | 1% | 75% | 3 | Search N+SUCCINYLAMINO+ACID+RACEMASE | Search N+SUCCINYLAMINO+ACID+RACEMASE |
| 8 | PSEUDOMONAS PUTIDA ATCC 12633 | 226745 | 1% | 75% | 3 | Search PSEUDOMONAS+PUTIDA+ATCC+12633 | Search PSEUDOMONAS+PUTIDA+ATCC+12633 |
| 9 | L RHAMNONATE | 201553 | 1% | 100% | 2 | Search L+RHAMNONATE | Search L+RHAMNONATE |
| 10 | MESO GALACTARIC ACID | 201553 | 1% | 100% | 2 | Search MESO+GALACTARIC+ACID | Search MESO+GALACTARIC+ACID |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | GERLT, JA , BABBITT, PC , RAYMENT, I , (2005) DIVERGENT EVOLUTION IN THE ENOLASE SUPERFAMILY: THE INTERPLAY OF MECHANISM AND SPECIFICITY.ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS. VOL. 433. ISSUE 1. P. 59-70 | 37 | 77% | 117 |
| 2 | GERLT, JA , BABBITT, PC , JACOBSON, MP , ALMO, SC , (2012) DIVERGENT EVOLUTION IN ENOLASE SUPERFAMILY: STRATEGIES FOR ASSIGNING FUNCTIONS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 287. ISSUE 1. P. 29-34 | 25 | 74% | 52 |
| 3 | GERLT, JA , BOUVIER, JT , DAVIDSON, DB , IMKER, HJ , SADKHIN, B , SLATER, DR , WHALEN, KL , (2015) ENZYME FUNCTION INITIATIVE-ENZYME SIMILARITY TOOL (EFI-EST): A WEB TOOL FOR GENERATING PROTEIN SEQUENCE SIMILARITY NETWORKS.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1854. ISSUE 8. P. 1019 -1037 | 24 | 48% | 27 |
| 4 | SWAMINATHAN, S , BURLEY, SK , ALMO, SC , GLASNER, ME , ODOKONYERO, D , RAGUMANI, S , LOPEZ, MS , BONANNO, JB , OZEROVA, NDS , WOODARD, DR , ET AL (2013) DIVERGENT EVOLUTION OF LIGAND BINDING IN THE O-SUCCINYLBENZOATE SYNTHASE FAMILY.BIOCHEMISTRY. VOL. 52. ISSUE 42. P. 7512 -7521 | 25 | 58% | 6 |
| 5 | MCMILLAN, AW , LOPEZ, MS , ZHU, MZ , MORSE, BC , YEO, IC , AMOS, J , HULL, K , ROMO, D , GLASNER, ME , (2014) ROLE OF AN ACTIVE SITE LOOP IN THE PROMISCUOUS ACTIVITIES OF AMYCOLATOPSIS SP T-1-60 NSAR/OSBS.BIOCHEMISTRY. VOL. 53. ISSUE 27. P. 4434 -4444 | 29 | 48% | 4 |
| 6 | SORIANO-MALDONADO, P , ANDUJAR-SANCHEZ, M , CLEMENTE-JIMENEZ, JM , RODRIGUEZ-VICO, F , HERAS-VAZQUEZ, FJL , MARTINEZ-RODRIGUEZ, S , (2015) BIOCHEMICAL AND MUTATIONAL CHARACTERIZATION OF N-SUCCINYL-AMINO ACID RACEMASE FROM GEOBACILLUS STEAROTHERMOPHILUS CECT49.MOLECULAR BIOTECHNOLOGY. VOL. 57. ISSUE 5. P. 454 -465 | 22 | 63% | 0 |
| 7 | ODOKONYERO, D , SAKAI, A , PATSKOVSKY, Y , MALASHKEVICH, VN , FEDOROV, AA , BONANNO, JB , FEDOROV, EV , TORO, R , AGARWAL, R , WANG, CX , ET AL (2014) LOSS OF QUATERNARY STRUCTURE IS ASSOCIATED WITH RAPID SEQUENCE DIVERGENCE IN THE OSBS FAMILY.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. VOL. 111. ISSUE 23. P. 8535-8540 | 28 | 47% | 1 |
| 8 | NAGAR, M , WYATT, BN , ST MAURICE, M , BEARNE, SL , (2015) INACTIVATION OF MANDE LATE RACEMASE BY 3-HYDROXYPYRUVATE REVEALS A POTENTIAL MECHANISTIC LINK BETWEEN ENZYME SUPERFAMILIES.BIOCHEMISTRY. VOL. 54. ISSUE 17. P. 2747 -2757 | 25 | 45% | 0 |
| 9 | RAKUS, JF , FEDOROV, AA , FEDOROV, EV , GLASNER, ME , VICK, JE , BABBITT, PC , ALMO, SC , GERLT, JA , (2007) EVOLUTION OF ENZYMATIC ACTIVITIES IN THE ENOLASE SUPERFAMILY: D-MANNONATE DEHYDRATASE FROM NOVOSPHINGOBIUM AROMATICIVORANS.BIOCHEMISTRY. VOL. 46. ISSUE 45. P. 12896-12908 | 21 | 62% | 10 |
| 10 | NAGAR, M , BEARNE, SL , (2015) AN ADDITIONAL ROLE FOR THE BRONSTED ACID-BASE CATALYSTS OF MANDELATE RACEMASE IN TRANSITION STATE STABILIZATION.BIOCHEMISTRY. VOL. 54. ISSUE 44. P. 6743 -6752 | 23 | 46% | 1 |
Classes with closest relation at Level 1 |