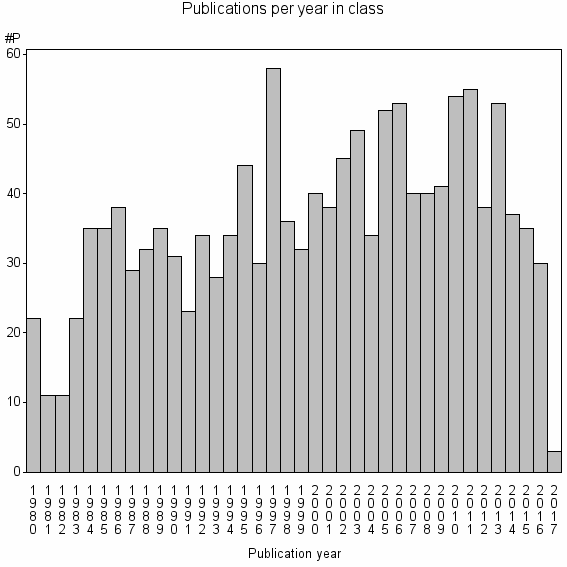
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 7535 | 1357 | 37.3 | 80% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 1115 | 2 | GENOME EDITING//CRISPR CAS9//CRISPR | 9706 |
| 7535 | 1 | SITE SPECIFIC RECOMBINATION//SERINE RECOMBINASE//PHIC31 INTEGRASE | 1357 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SITE SPECIFIC RECOMBINATION | authKW | 1464779 | 13% | 37% | 174 |
| 2 | SERINE RECOMBINASE | authKW | 529117 | 2% | 87% | 27 |
| 3 | PHIC31 INTEGRASE | authKW | 281241 | 1% | 63% | 20 |
| 4 | PHI C31 INTEGRASE | authKW | 238885 | 1% | 56% | 19 |
| 5 | TYROSINE RECOMBINASE | authKW | 206699 | 2% | 44% | 21 |
| 6 | SITE SPECIFIC DNA RECOMBINATION | authKW | 202498 | 1% | 75% | 12 |
| 7 | LAMBDA INTEGRASE | authKW | 190585 | 1% | 71% | 12 |
| 8 | RECOMBINASE MEDIATED CASSETTE EXCHANGE | authKW | 170521 | 1% | 63% | 12 |
| 9 | PHAGE INTEGRASE | authKW | 165682 | 1% | 82% | 9 |
| 10 | BACTERIOPHAGE HK022 | authKW | 144000 | 1% | 80% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 8261 | 63% | 0% | 861 |
| 2 | Genetics & Heredity | 1683 | 16% | 0% | 213 |
| 3 | Microbiology | 1166 | 13% | 0% | 182 |
| 4 | Cell Biology | 1135 | 16% | 0% | 222 |
| 5 | Biotechnology & Applied Microbiology | 414 | 9% | 0% | 119 |
| 6 | Developmental Biology | 278 | 3% | 0% | 44 |
| 7 | Biology | 276 | 5% | 0% | 64 |
| 8 | Biochemical Research Methods | 180 | 5% | 0% | 65 |
| 9 | Biophysics | 71 | 4% | 0% | 51 |
| 10 | Medicine, Research & Experimental | 49 | 3% | 0% | 47 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ANDERSONS BIOMED LIFE SCI | 22500 | 0% | 100% | 1 |
| 2 | ANIM DIS MODELING | 22500 | 0% | 100% | 1 |
| 3 | AUSTRALIAN BACTERIAL PATHOGENESIS PROGRAMME | 22500 | 0% | 100% | 1 |
| 4 | BACTERIAL GENOME PLASTIC UNIT | 22500 | 0% | 100% | 1 |
| 5 | BIOCHEM MOLL BIOL GRP | 22500 | 0% | 100% | 1 |
| 6 | BIOL MED BOX G | 22500 | 0% | 100% | 1 |
| 7 | BIOL SCI IFM | 22500 | 0% | 100% | 1 |
| 8 | BIOMED LIFE | 22500 | 0% | 100% | 1 |
| 9 | CELL BIOL GS10 | 22500 | 0% | 100% | 1 |
| 10 | CHIRURG FOR UNGS OR | 22500 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF MOLECULAR BIOLOGY | 20523 | 11% | 1% | 146 |
| 2 | NUCLEIC ACIDS RESEARCH | 8648 | 9% | 0% | 124 |
| 3 | COLD SPRING HARBOR SYMPOSIA ON QUANTITATIVE BIOLOGY | 5336 | 2% | 1% | 23 |
| 4 | GENESIS | 4799 | 1% | 1% | 18 |
| 5 | FLY | 4374 | 1% | 2% | 9 |
| 6 | JOURNAL OF BACTERIOLOGY | 4352 | 6% | 0% | 81 |
| 7 | MOLECULAR MICROBIOLOGY | 3944 | 3% | 0% | 46 |
| 8 | EMBO JOURNAL | 3354 | 4% | 0% | 51 |
| 9 | CELL | 2852 | 3% | 0% | 44 |
| 10 | JOURNAL OF KNOT THEORY AND ITS RAMIFICATIONS | 2545 | 1% | 1% | 14 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SITE SPECIFIC RECOMBINATION | 1464779 | 13% | 37% | 174 | Search SITE+SPECIFIC+RECOMBINATION | Search SITE+SPECIFIC+RECOMBINATION |
| 2 | SERINE RECOMBINASE | 529117 | 2% | 87% | 27 | Search SERINE+RECOMBINASE | Search SERINE+RECOMBINASE |
| 3 | PHIC31 INTEGRASE | 281241 | 1% | 63% | 20 | Search PHIC31+INTEGRASE | Search PHIC31+INTEGRASE |
| 4 | PHI C31 INTEGRASE | 238885 | 1% | 56% | 19 | Search PHI+C31+INTEGRASE | Search PHI+C31+INTEGRASE |
| 5 | TYROSINE RECOMBINASE | 206699 | 2% | 44% | 21 | Search TYROSINE+RECOMBINASE | Search TYROSINE+RECOMBINASE |
| 6 | SITE SPECIFIC DNA RECOMBINATION | 202498 | 1% | 75% | 12 | Search SITE+SPECIFIC+DNA+RECOMBINATION | Search SITE+SPECIFIC+DNA+RECOMBINATION |
| 7 | LAMBDA INTEGRASE | 190585 | 1% | 71% | 12 | Search LAMBDA+INTEGRASE | Search LAMBDA+INTEGRASE |
| 8 | RECOMBINASE MEDIATED CASSETTE EXCHANGE | 170521 | 1% | 63% | 12 | Search RECOMBINASE+MEDIATED+CASSETTE+EXCHANGE | Search RECOMBINASE+MEDIATED+CASSETTE+EXCHANGE |
| 9 | PHAGE INTEGRASE | 165682 | 1% | 82% | 9 | Search PHAGE+INTEGRASE | Search PHAGE+INTEGRASE |
| 10 | BACTERIOPHAGE HK022 | 144000 | 1% | 80% | 8 | Search BACTERIOPHAGE+HK022 | Search BACTERIOPHAGE+HK022 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | GRINDLEY, NDF , WHITESON, KL , RICE, PA , (2006) MECHANISMS OF SITE-SPECIFIC RECOMBINATION.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 75. ISSUE . P. 567 -605 | 106 | 79% | 307 |
| 2 | LEE, L , SADOWSKI, PD , (2005) STRAND SELECTION BY THE TYROSINE RECOMBINASES.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 80. VOL. 80. ISSUE . P. 1 -42 | 116 | 78% | 10 |
| 3 | GROTH, AC , CALOS, MP , (2004) PHAGE INTEGRASES: BIOLOGY AND APPLICATIONS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 335. ISSUE 3. P. 667-678 | 75 | 85% | 204 |
| 4 | FOGG, PCM , COLLOMS, S , ROSSER, S , STARK, M , SMITH, MCM , (2014) NEW APPLICATIONS FOR PHAGE INTEGRASES.JOURNAL OF MOLECULAR BIOLOGY. VOL. 426. ISSUE 15. P. 2703 -2716 | 79 | 65% | 21 |
| 5 | MA, CH , LIU, YT , SAVVA, CG , ROWLEY, PA , CANNON, B , FAN, HF , RUSSELL, R , HOLZENBURG, A , JAYARAM, M , (2014) ORGANIZATION OF DNA PARTNERS AND STRAND EXCHANGE MECHANISMS DURING FLP SITE-SPECIFIC RECOMBINATION ANALYZED BY DIFFERENCE TOPOLOGY, SINGLE MOLECULE FRET AND SINGLE MOLECULE TPM.JOURNAL OF MOLECULAR BIOLOGY. VOL. 426. ISSUE 4. P. 793 -815 | 56 | 86% | 2 |
| 6 | GAJ, T , SIRK, SJ , BARBAS, CF , (2014) EXPANDING THE SCOPE OF SITE-SPECIFIC RECOMBINASES FOR GENETIC AND METABOLIC ENGINEERING.BIOTECHNOLOGY AND BIOENGINEERING. VOL. 111. ISSUE 1. P. 1-15 | 76 | 52% | 17 |
| 7 | OLORUNNIJI, FJ , ROSSER, SJ , STARK, WM , (2016) SITE-SPECIFIC RECOMBINASES: MOLECULAR MACHINES FOR THE GENETIC REVOLUTION.BIOCHEMICAL JOURNAL. VOL. 473. ISSUE . P. 673 -684 | 65 | 52% | 3 |
| 8 | RAJEEV, L , MALANOWSKA, K , GARDNER, JF , (2009) CHALLENGING A PARADIGM: THE ROLE OF DNA HOMOLOGY IN TYROSINE RECOMBINASE REACTIONS.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 73. ISSUE 2. P. 300 -+ | 59 | 75% | 31 |
| 9 | BOWYER, J , ZHAO, J , SUBSOONTORN, P , WONG, W , ROSSER, S , BATES, D , (2016) MECHANISTIC MODELING OF A REWRITABLE RECOMBINASE ADDRESSABLE DATA MODULE.IEEE TRANSACTIONS ON BIOMEDICAL CIRCUITS AND SYSTEMS. VOL. 10. ISSUE 6. P. 1161 -1170 | 45 | 83% | 1 |
| 10 | VAN DUYNE, GD , RUTHERFORD, K , (2013) LARGE SERINE RECOMBINASE DOMAIN STRUCTURE AND ATTACHMENT SITE BINDING.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 48. ISSUE 5. P. 476-491 | 51 | 78% | 10 |
Classes with closest relation at Level 1 |