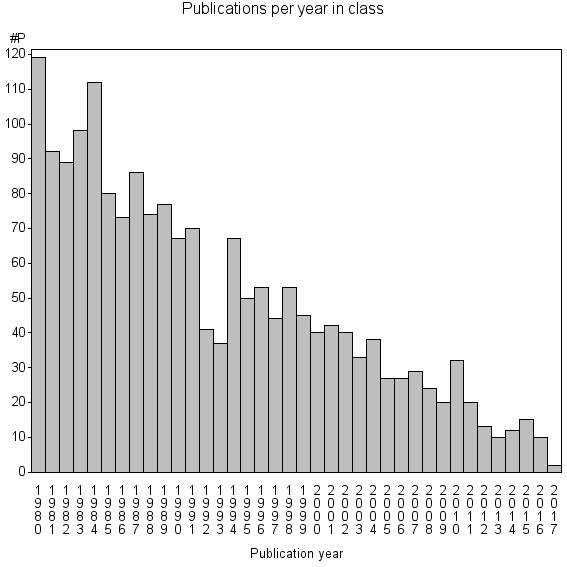
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 4021 | 1861 | 36.5 | 60% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 3063 | 2 | DNA TRANSPOSITION//PHAGE MU//TRANSPOSASE | 2429 |
| 4021 | 1 | DNA TRANSPOSITION//PHAGE MU//TRANSPOSASE | 1861 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA TRANSPOSITION | authKW | 582836 | 2% | 79% | 45 |
| 2 | PHAGE MU | authKW | 425377 | 2% | 79% | 33 |
| 3 | TRANSPOSASE | authKW | 238791 | 3% | 23% | 62 |
| 4 | BACTERIOPHAGE MU | authKW | 234364 | 1% | 71% | 20 |
| 5 | TN10 | authKW | 211680 | 1% | 65% | 20 |
| 6 | TRANSPOSOSOME | authKW | 196875 | 1% | 100% | 12 |
| 7 | TRANSPOSITION | authKW | 166297 | 5% | 12% | 86 |
| 8 | IS911 | authKW | 147657 | 0% | 100% | 9 |
| 9 | MUA TRANSPOSASE | authKW | 147657 | 0% | 100% | 9 |
| 10 | MICROBIOL GENET MOL | address | 115781 | 3% | 13% | 56 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 9683 | 30% | 0% | 567 |
| 2 | Microbiology | 8229 | 28% | 0% | 528 |
| 3 | Biochemistry & Molecular Biology | 6527 | 49% | 0% | 921 |
| 4 | Cell Biology | 1081 | 14% | 0% | 261 |
| 5 | Biology | 784 | 6% | 0% | 120 |
| 6 | Virology | 174 | 3% | 0% | 53 |
| 7 | Evolutionary Biology | 61 | 2% | 0% | 30 |
| 8 | Biotechnology & Applied Microbiology | 38 | 3% | 0% | 65 |
| 9 | Developmental Biology | 8 | 1% | 0% | 15 |
| 10 | Biochemical Research Methods | 6 | 2% | 0% | 30 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MICROBIOL GENET MOL | 115781 | 3% | 13% | 56 |
| 2 | UMR5100 | 60492 | 1% | 25% | 15 |
| 3 | PROGRAM CELLULAR BIOTECHNOL | 34637 | 1% | 16% | 13 |
| 4 | MICROORGANISMS GENET ERS | 32813 | 0% | 100% | 2 |
| 5 | UPR 9007 | 24120 | 0% | 29% | 5 |
| 6 | ACIDI NUCLE | 16406 | 0% | 100% | 1 |
| 7 | BASS MOL BIOPHYS BIOCHEM | 16406 | 0% | 100% | 1 |
| 8 | BIO MICROBIOL KLINGELBERGSTR 70 | 16406 | 0% | 100% | 1 |
| 9 | BIOL SCIMARINE ENVIRONM BIOL | 16406 | 0% | 100% | 1 |
| 10 | BIOQUIM POLIGONO CAZONA SN | 16406 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND GENERAL GENETICS | 32724 | 5% | 2% | 98 |
| 2 | PLASMID | 31070 | 3% | 3% | 62 |
| 3 | JOURNAL OF BACTERIOLOGY | 21910 | 11% | 1% | 211 |
| 4 | COLD SPRING HARBOR SYMPOSIA ON QUANTITATIVE BIOLOGY | 16995 | 3% | 2% | 48 |
| 5 | GENE | 13764 | 7% | 1% | 128 |
| 6 | MOLECULAR & GENERAL GENETICS | 9197 | 2% | 2% | 37 |
| 7 | JOURNAL OF MOLECULAR BIOLOGY | 8867 | 6% | 0% | 113 |
| 8 | MOBILE DNA | 8669 | 0% | 6% | 9 |
| 9 | GENETIKA | 7186 | 2% | 1% | 44 |
| 10 | MOLECULAR MICROBIOLOGY | 6109 | 4% | 1% | 67 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA TRANSPOSITION | 582836 | 2% | 79% | 45 | Search DNA+TRANSPOSITION | Search DNA+TRANSPOSITION |
| 2 | PHAGE MU | 425377 | 2% | 79% | 33 | Search PHAGE+MU | Search PHAGE+MU |
| 3 | TRANSPOSASE | 238791 | 3% | 23% | 62 | Search TRANSPOSASE | Search TRANSPOSASE |
| 4 | BACTERIOPHAGE MU | 234364 | 1% | 71% | 20 | Search BACTERIOPHAGE+MU | Search BACTERIOPHAGE+MU |
| 5 | TN10 | 211680 | 1% | 65% | 20 | Search TN10 | Search TN10 |
| 6 | TRANSPOSOSOME | 196875 | 1% | 100% | 12 | Search TRANSPOSOSOME | Search TRANSPOSOSOME |
| 7 | TRANSPOSITION | 166297 | 5% | 12% | 86 | Search TRANSPOSITION | Search TRANSPOSITION |
| 8 | IS911 | 147657 | 0% | 100% | 9 | Search IS911 | Search IS911 |
| 9 | MUA TRANSPOSASE | 147657 | 0% | 100% | 9 | Search MUA+TRANSPOSASE | Search MUA+TRANSPOSASE |
| 10 | INSERTION SEQUENCE | 100952 | 2% | 15% | 41 | Search INSERTION+SEQUENCE | Search INSERTION+SEQUENCE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MAHILLON, J , CHANDLER, M , (1998) INSERTION SEQUENCES.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 62. ISSUE 3. P. 725 -+ | 180 | 53% | 896 |
| 2 | MITKINA, LN , (2003) TRANSPOSITION AS A WAY OF EXISTENCE: PHAGE MU.RUSSIAN JOURNAL OF GENETICS. VOL. 39. ISSUE 5. P. 519-536 | 100 | 78% | 0 |
| 3 | NAGY, Z , CHANDLER, M , (2004) REGULATION OF TRANSPOSITION IN BACTERIA.RESEARCH IN MICROBIOLOGY. VOL. 155. ISSUE 5. P. 387-398 | 71 | 80% | 98 |
| 4 | REZNIKOFF, WS , (2008) TRANSPOSON TN5.ANNUAL REVIEW OF GENETICS. VOL. 42. ISSUE . P. 269-286 | 56 | 92% | 42 |
| 5 | LEWIS, LA , ASTATKE, M , UMEKUBO, PT , ALVI, S , SABY, R , AFROSE, J , OLIVEIRA, PH , MONTEIRO, GA , PRAZERES, DMF , (2012) PROTEIN-DNA INTERACTIONS DEFINE THE MECHANISTIC ASPECTS OF CIRCLE FORMATION AND INSERTION REACTIONS IN IS2 TRANSPOSITION.MOBILE DNA. VOL. 3. ISSUE . P. - | 59 | 86% | 0 |
| 6 | CHOI, W , SAHA, RP , JANG, S , HARSHEY, RM , (2014) CONTROLLING DNA DEGRADATION FROM A DISTANCE: A NEW ROLE FOR THE MU TRANSPOSITION ENHANCER.MOLECULAR MICROBIOLOGY. VOL. 94. ISSUE 3. P. 595 -608 | 53 | 80% | 0 |
| 7 | SIGUIER, P , GOURBEYRE, E , CHANDLER, M , (2014) BACTERIAL INSERTION SEQUENCES: THEIR GENOMIC IMPACT AND DIVERSITY.FEMS MICROBIOLOGY REVIEWS. VOL. 38. ISSUE 5. P. 865 -891 | 74 | 38% | 42 |
| 8 | KLECKNER, N , (1990) REGULATION OF TRANSPOSITION IN BACTERIA.ANNUAL REVIEW OF CELL BIOLOGY. VOL. 6. ISSUE . P. 297-327 | 89 | 85% | 62 |
| 9 | KLECKNER, N , CHALMERS, RM , KWON, D , SAKAI, J , BOLLAND, S , (1996) TN10 AND IS10 TRANSPOSITION AND CHROMOSOME REARRANGEMENTS: MECHANISM AND REGULATION IN VIVO AND IN VITRO.TRANSPOSABLE ELEMENTS. VOL. 204. ISSUE . P. 49 -82 | 66 | 87% | 64 |
| 10 | HAREN, L , TON-HOANG, B , CHANDLER, M , (1999) INTEGRATING DNA: TRANSPOSASES AND RETROVIRAL INTEGRASES.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 53. ISSUE . P. 245-281 | 88 | 51% | 159 |
Classes with closest relation at Level 1 |