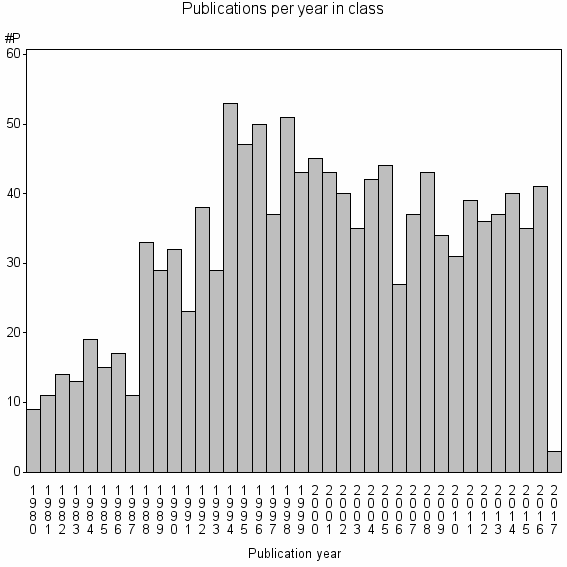
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8709 | 1226 | 44.1 | 80% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 371 | 2 | RNA POLYMERASE//HFQ//JOURNAL OF BACTERIOLOGY | 17271 |
| 8709 | 1 | H NS//HISTONE LIKE PROTEIN//NUCLEOID ASSOCIATED PROTEIN | 1226 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | H NS | authKW | 852971 | 6% | 50% | 68 |
| 2 | HISTONE LIKE PROTEIN | authKW | 720401 | 4% | 59% | 49 |
| 3 | NUCLEOID ASSOCIATED PROTEIN | authKW | 355968 | 2% | 53% | 27 |
| 4 | HU PROTEIN | authKW | 310010 | 2% | 66% | 19 |
| 5 | INTEGRATION HOST FACTOR | authKW | 300610 | 2% | 46% | 26 |
| 6 | BACTERIAL CHROMATIN | authKW | 249049 | 1% | 100% | 10 |
| 7 | PHYSIOL BACTERIENNE | address | 247578 | 1% | 76% | 13 |
| 8 | HHA | authKW | 203378 | 1% | 58% | 14 |
| 9 | IHF | authKW | 196477 | 2% | 33% | 24 |
| 10 | HU | authKW | 195184 | 2% | 29% | 27 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Microbiology | 7789 | 34% | 0% | 413 |
| 2 | Biochemistry & Molecular Biology | 7487 | 64% | 0% | 779 |
| 3 | Biophysics | 1155 | 12% | 0% | 147 |
| 4 | Genetics & Heredity | 337 | 8% | 0% | 101 |
| 5 | Cell Biology | 277 | 10% | 0% | 118 |
| 6 | CYTOLOGY & HISTOLOGY | 45 | 0% | 0% | 4 |
| 7 | Biology | 37 | 2% | 0% | 28 |
| 8 | Biochemical Research Methods | 23 | 2% | 0% | 30 |
| 9 | Biotechnology & Applied Microbiology | 7 | 3% | 0% | 32 |
| 10 | Crystallography | 6 | 1% | 0% | 16 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PHYSIOL BACTERIENNE | 247578 | 1% | 76% | 13 |
| 2 | LEIDEN CHEM CELL OBSERV | 174334 | 1% | 100% | 7 |
| 3 | PROGRAM BIOTECHNOL BIOENGN | 68961 | 0% | 46% | 6 |
| 4 | CNRS URA 409 | 49810 | 0% | 100% | 2 |
| 5 | PROGRAM BIOTECHNOL SCI ENGN | 49810 | 0% | 100% | 2 |
| 6 | CELL OBSERV | 44826 | 0% | 60% | 3 |
| 7 | MOYNE PREVENT MED | 37865 | 2% | 7% | 22 |
| 8 | FRE2326 | 33205 | 0% | 67% | 2 |
| 9 | PLASMA MEMBRANE NUCL SIGNALING | 28803 | 1% | 13% | 9 |
| 10 | AGR GENETHIGASHI KU | 24905 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR MICROBIOLOGY | 14029 | 7% | 1% | 82 |
| 2 | JOURNAL OF BACTERIOLOGY | 12805 | 11% | 0% | 131 |
| 3 | JOURNAL OF MOLECULAR BIOLOGY | 6758 | 7% | 0% | 80 |
| 4 | BIOCHIMIE | 2992 | 2% | 0% | 27 |
| 5 | EXTREMOPHILES | 2955 | 1% | 1% | 13 |
| 6 | NUCLEIC ACIDS RESEARCH | 2417 | 5% | 0% | 63 |
| 7 | CURRENT OPINION IN MICROBIOLOGY | 1941 | 1% | 1% | 12 |
| 8 | PLASMID | 1469 | 1% | 1% | 11 |
| 9 | FEMS MICROBIOLOGY LETTERS | 1187 | 2% | 0% | 28 |
| 10 | BIOCHEMISTRY | 1140 | 4% | 0% | 51 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | H NS | 852971 | 6% | 50% | 68 | Search H+NS | Search H+NS |
| 2 | HISTONE LIKE PROTEIN | 720401 | 4% | 59% | 49 | Search HISTONE+LIKE+PROTEIN | Search HISTONE+LIKE+PROTEIN |
| 3 | NUCLEOID ASSOCIATED PROTEIN | 355968 | 2% | 53% | 27 | Search NUCLEOID+ASSOCIATED+PROTEIN | Search NUCLEOID+ASSOCIATED+PROTEIN |
| 4 | HU PROTEIN | 310010 | 2% | 66% | 19 | Search HU+PROTEIN | Search HU+PROTEIN |
| 5 | INTEGRATION HOST FACTOR | 300610 | 2% | 46% | 26 | Search INTEGRATION+HOST+FACTOR | Search INTEGRATION+HOST+FACTOR |
| 6 | BACTERIAL CHROMATIN | 249049 | 1% | 100% | 10 | Search BACTERIAL+CHROMATIN | Search BACTERIAL+CHROMATIN |
| 7 | HHA | 203378 | 1% | 58% | 14 | Search HHA | Search HHA |
| 8 | IHF | 196477 | 2% | 33% | 24 | Search IHF | Search IHF |
| 9 | HU | 195184 | 2% | 29% | 27 | Search HU | Search HU |
| 10 | H NS PROTEIN | 149429 | 0% | 100% | 6 | Search H+NS+PROTEIN | Search H+NS+PROTEIN |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | GROVE, A , (2011) FUNCTIONAL EVOLUTION OF BACTERIAL HISTONE-LIKE HU PROTEINS.CURRENT ISSUES IN MOLECULAR BIOLOGY. VOL. 13. ISSUE 1. P. 1 -11 | 75 | 72% | 36 |
| 2 | DORMAN, CJ , (2014) H-NS-LIKE NUCLEOID-ASSOCIATED PROTEINS, MOBILE GENETIC ELEMENTS AND HORIZONTAL GENE TRANSFER IN BACTERIA.PLASMID. VOL. 75. ISSUE . P. 1 -11 | 76 | 53% | 17 |
| 3 | DORMAN, CJ , (2004) H-NS: A UNIVERSAL REGULATOR FOR A DYNAMIC GENOME.NATURE REVIEWS MICROBIOLOGY. VOL. 2. ISSUE 5. P. 391-400 | 66 | 63% | 302 |
| 4 | SWINGER, KK , RICE, PA , (2004) IHF AND HU: FLEXIBLE ARCHITECTS OF BENT DNA.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 14. ISSUE 1. P. 28-35 | 51 | 85% | 188 |
| 5 | ZHANG, ZF , GUO, L , HUANG, L , (2012) ARCHAEAL CHROMATIN PROTEINS.SCIENCE CHINA-LIFE SCIENCES. VOL. 55. ISSUE 5. P. 377-385 | 51 | 82% | 10 |
| 6 | WINARDHI, RS , YAN, J , KENNEY, LJ , (2015) H-NS REGULATES GENE EXPRESSION AND COMPACTS THE NUCLEOID: INSIGHTS FROM SINGLE-MOLECULE EXPERIMENTS.BIOPHYSICAL JOURNAL. VOL. 109. ISSUE 7. P. 1321 -1329 | 47 | 76% | 6 |
| 7 | SINGH, K , MILSTEIN, JN , NAVARRE, WW , (2016) XENOGENEIC SILENCING AND ITS IMPACT ON BACTERIAL GENOMES.ANNUAL REVIEW OF MICROBIOLOGY, VOL 70. VOL. 70. ISSUE . P. 199 -213 | 55 | 65% | 1 |
| 8 | NAVARRE, WW , MCCLELLAND, M , LIBBY, SJ , FANG, FC , (2007) SILENCING OF XENOGENEIC DNA BY H-NS - FACILITATION OF LATERAL GENE TRANSFER IN BACTERIA BY A DEFENSE SYSTEM THAT RECOGNIZES FOREIGN DNA.GENES & DEVELOPMENT. VOL. 21. ISSUE 12. P. 1456 -1471 | 82 | 46% | 123 |
| 9 | SHINTANI, M , SUZUKI-MINAKUCHI, C , NOJIRI, H , (2015) NUCLEOID-ASSOCIATED PROTEINS ENCODED ON PLASMIDS: OCCURRENCE AND MODE OF FUNCTION.PLASMID. VOL. 80. ISSUE . P. 32 -44 | 61 | 59% | 4 |
| 10 | LANDICK, R , WADE, JT , GRAINGER, DC , (2015) H-NS AND RNA POLYMERASE: A LOVE-HATE RELATIONSHIP?.CURRENT OPINION IN MICROBIOLOGY. VOL. 24. ISSUE . P. 53 -59 | 42 | 76% | 6 |
Classes with closest relation at Level 1 |