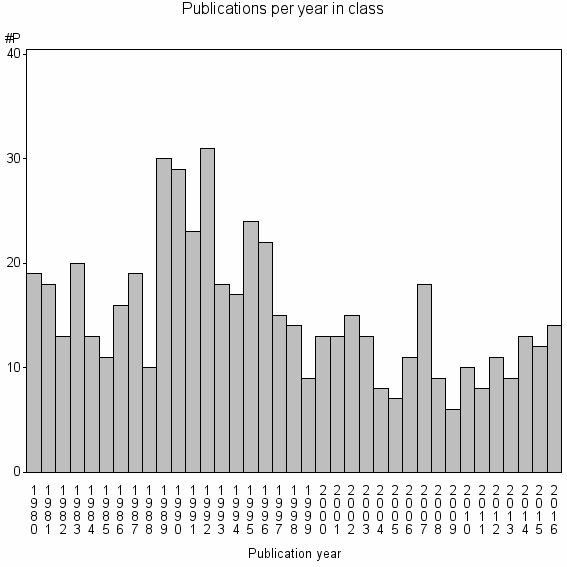
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 18221 | 561 | 37.2 | 60% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 959 | 2 | BACTERIOPHAGE//PHAGE THERAPY//ENDOLYSIN | 10780 |
| 18221 | 1 | SKIRBALL PROGRAM MOL PATHOGENESIS//RETRON//MSDNA | 561 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SKIRBALL PROGRAM MOL PATHOGENESIS | address | 583160 | 3% | 71% | 15 |
| 2 | RETRON | authKW | 340174 | 2% | 63% | 10 |
| 3 | MSDNA | authKW | 244928 | 1% | 75% | 6 |
| 4 | MOLECULAR PIRACY | authKW | 178126 | 1% | 55% | 6 |
| 5 | BACTERIOPHAGE P2 | authKW | 170087 | 1% | 63% | 5 |
| 6 | PHAGE P4 | authKW | 163287 | 1% | 100% | 3 |
| 7 | BACTERIOPHAGE N15 | authKW | 145141 | 1% | 67% | 4 |
| 8 | BACTERIOPHAGE P2 P4 | authKW | 108858 | 0% | 100% | 2 |
| 9 | MULTICOPY SINGLE STRANDED DNA | authKW | 108858 | 0% | 100% | 2 |
| 10 | PROPHAGE IMMUNITY | authKW | 108858 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Microbiology | 4490 | 38% | 0% | 211 |
| 2 | Virology | 3367 | 19% | 0% | 106 |
| 3 | Biochemistry & Molecular Biology | 1413 | 43% | 0% | 240 |
| 4 | Genetics & Heredity | 767 | 16% | 0% | 92 |
| 5 | Cell Biology | 17 | 5% | 0% | 28 |
| 6 | Biotechnology & Applied Microbiology | 15 | 4% | 0% | 21 |
| 7 | Biochemical Research Methods | 1 | 1% | 0% | 8 |
| 8 | Mathematical & Computational Biology | 1 | 1% | 0% | 3 |
| 9 | Biology | 1 | 1% | 0% | 6 |
| 10 | Biophysics | 0 | 1% | 0% | 7 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SKIRBALL PROGRAM MOL PATHOGENESIS | 583160 | 3% | 71% | 15 |
| 2 | QUIM BIOQUIM BIOL MOL | 89211 | 2% | 16% | 10 |
| 3 | BIOL BIOTEKNOL OSLO | 54429 | 0% | 100% | 1 |
| 4 | BIOL STRUCT BIOCOMP PROGRAMMING | 54429 | 0% | 100% | 1 |
| 5 | DIPARTIMENTO GENET BIOL MICROORGANISM | 54429 | 0% | 100% | 1 |
| 6 | INTEGRAT OM GRP | 54429 | 0% | 100% | 1 |
| 7 | JET PROP 125224 | 54429 | 0% | 100% | 1 |
| 8 | KAROLINSKA IMAGING | 54429 | 0% | 100% | 1 |
| 9 | MICRBIAL BIOCHEM | 54429 | 0% | 100% | 1 |
| 10 | MOLEC CELL BIOL 401 BARKER HALL | 54429 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BACTERIOLOGY | 11852 | 15% | 0% | 85 |
| 2 | VIROLOGY | 10181 | 11% | 0% | 59 |
| 3 | MOLECULAR MICROBIOLOGY | 8053 | 7% | 0% | 42 |
| 4 | JOURNAL OF THE AMERICAN MEDICAL TECHNOLOGISTS | 6046 | 0% | 11% | 1 |
| 5 | JOURNAL OF MOLECULAR BIOLOGY | 4282 | 8% | 0% | 43 |
| 6 | PLASMID | 3852 | 2% | 1% | 12 |
| 7 | MOLECULAR AND GENERAL GENETICS | 3648 | 3% | 0% | 18 |
| 8 | GENE | 2170 | 5% | 0% | 28 |
| 9 | MOLECULAR & GENERAL GENETICS | 1802 | 2% | 0% | 9 |
| 10 | JOURNAL OF VIROLOGY | 891 | 5% | 0% | 26 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RETRON | 340174 | 2% | 63% | 10 | Search RETRON | Search RETRON |
| 2 | MSDNA | 244928 | 1% | 75% | 6 | Search MSDNA | Search MSDNA |
| 3 | MOLECULAR PIRACY | 178126 | 1% | 55% | 6 | Search MOLECULAR+PIRACY | Search MOLECULAR+PIRACY |
| 4 | BACTERIOPHAGE P2 | 170087 | 1% | 63% | 5 | Search BACTERIOPHAGE+P2 | Search BACTERIOPHAGE+P2 |
| 5 | PHAGE P4 | 163287 | 1% | 100% | 3 | Search PHAGE+P4 | Search PHAGE+P4 |
| 6 | BACTERIOPHAGE N15 | 145141 | 1% | 67% | 4 | Search BACTERIOPHAGE+N15 | Search BACTERIOPHAGE+N15 |
| 7 | BACTERIOPHAGE P2 P4 | 108858 | 0% | 100% | 2 | Search BACTERIOPHAGE+P2+P4 | Search BACTERIOPHAGE+P2+P4 |
| 8 | MULTICOPY SINGLE STRANDED DNA | 108858 | 0% | 100% | 2 | Search MULTICOPY+SINGLE+STRANDED+DNA | Search MULTICOPY+SINGLE+STRANDED+DNA |
| 9 | PROPHAGE IMMUNITY | 108858 | 0% | 100% | 2 | Search PROPHAGE+IMMUNITY | Search PROPHAGE+IMMUNITY |
| 10 | SAPI | 108854 | 1% | 50% | 4 | Search SAPI | Search SAPI |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LINDQVIST, BH , DEHO, G , CALENDAR, R , (1993) MECHANISMS OF GENOME PROPAGATION AND HELPER EXPLOITATION BY SATELLITE PHAGE-P4.MICROBIOLOGICAL REVIEWS. VOL. 57. ISSUE 3. P. 683 -702 | 93 | 71% | 83 |
| 2 | CHRISTIE, GE , DOKLAND, T , (2012) PIRATES OF THE CAUDOVIRALES.VIROLOGY. VOL. 434. ISSUE 2. P. 210-221 | 59 | 59% | 15 |
| 3 | LAMPSON, B , INOUYE, M , INOUYE, S , (2001) THE MSDNAS OF BACTERIA.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 67. VOL. 67. ISSUE . P. 65-91 | 51 | 84% | 13 |
| 4 | CARPENA, N , MANNING, KA , DOKLAND, T , MARINA, A , PENADES, JR , (2016) CONVERGENT EVOLUTION OF PATHOGENICITY ISLANDS IN HELPER COS PHAGE INTERFERENCE.PHILOSOPHICAL TRANSACTIONS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES. VOL. 371. ISSUE 1707. P. - | 33 | 75% | 1 |
| 5 | BRIANI, F , DEHO, G , FORTI, F , GHISOTTI, D , (2001) THE PLASMID STATUS OF SATELLITE BACTERIOPHAGE P4.PLASMID. VOL. 45. ISSUE 1. P. 1 -17 | 54 | 68% | 34 |
| 6 | LAMPSON, BC , INOUYE, M , INOUYE, S , (2005) RETRONS, MSDNA, AND THE BACTERIAL GENOME.CYTOGENETIC AND GENOME RESEARCH. VOL. 110. ISSUE 1-4. P. 491-499 | 42 | 69% | 31 |
| 7 | PENADES, JR , CHRISTIE, GE , (2015) THE PHAGE-INDUCIBLE CHROMOSOMAL ISLANDS: A FAMILY OF HIGHLY EVOLVED MOLECULAR PARASITES.ANNUAL REVIEW OF VIROLOGY, VOL 2. VOL. 2. ISSUE . P. 181 -201 | 37 | 57% | 4 |
| 8 | PORTELLI, R , DODD, IB , XUE, Q , EGAN, JB , (1998) THE LATE-EXPRESSED REGION OF THE TEMPERATE COLIPHAGE 186 GENOME.VIROLOGY. VOL. 248. ISSUE 1. P. 117-130 | 38 | 69% | 13 |
| 9 | CHRISTIE, GE , CALENDAR, R , (1990) INTERACTIONS BETWEEN SATELLITE BACTERIOPHAGE-P4 AND ITS HELPERS.ANNUAL REVIEW OF GENETICS. VOL. 24. ISSUE . P. 465-490 | 46 | 66% | 55 |
| 10 | NILSSON, AS , HAGGARD-LJUNGQUIST, E , (2007) EVOLUTION OF P2-LIKE PHAGES AND THEIR IMPACT ON BACTERIAL EVOLUTION.RESEARCH IN MICROBIOLOGY. VOL. 158. ISSUE 4. P. 311 -317 | 23 | 74% | 15 |
Classes with closest relation at Level 1 |