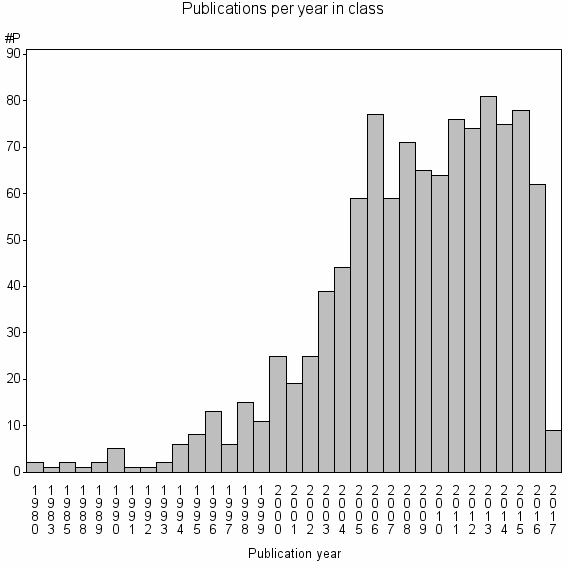
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 10256 | 1078 | 47.1 | 84% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 10256 | 1 | CODON MODELS//POSITIVE SELECTION//MOLECULAR BIOLOGY AND EVOLUTION | 1078 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CODON MODELS | authKW | 768093 | 4% | 68% | 40 |
| 2 | POSITIVE SELECTION | authKW | 482331 | 14% | 11% | 152 |
| 3 | MOLECULAR BIOLOGY AND EVOLUTION | journal | 276368 | 22% | 4% | 233 |
| 4 | CODON SUBSTITUTION MODEL | authKW | 241362 | 1% | 61% | 14 |
| 5 | DN DS | authKW | 207683 | 2% | 33% | 22 |
| 6 | BRANCH SITE MODEL | authKW | 181273 | 1% | 80% | 8 |
| 7 | PROTEIN EVOLUTION | authKW | 166836 | 6% | 9% | 66 |
| 8 | SYNONYMOUS SUBSTITUTION | authKW | 139021 | 2% | 27% | 18 |
| 9 | BRANCH SITE TEST | authKW | 113297 | 0% | 100% | 4 |
| 10 | CLADE MODEL | authKW | 113297 | 0% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Evolutionary Biology | 61953 | 50% | 0% | 538 |
| 2 | Genetics & Heredity | 26763 | 65% | 0% | 697 |
| 3 | Biochemistry & Molecular Biology | 4351 | 53% | 0% | 568 |
| 4 | Mathematical & Computational Biology | 3944 | 11% | 0% | 122 |
| 5 | Biochemical Research Methods | 758 | 10% | 0% | 104 |
| 6 | Biotechnology & Applied Microbiology | 691 | 12% | 0% | 129 |
| 7 | Biology | 312 | 5% | 0% | 59 |
| 8 | Statistics & Probability | 45 | 2% | 0% | 23 |
| 9 | Ecology | 23 | 3% | 0% | 30 |
| 10 | Biophysics | 10 | 2% | 0% | 25 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | URM 5506 | 84973 | 0% | 100% | 3 |
| 2 | BIOINFORMAT MOL EVOLUT GRP | 42478 | 1% | 25% | 6 |
| 3 | EA 3781 EVOLUT BIOL | 37764 | 0% | 67% | 2 |
| 4 | EHLTH INNOVAT PLATFORM | 37764 | 0% | 67% | 2 |
| 5 | ADV COMPUTAT SCI SYNTHET BIOL TEAM | 28324 | 0% | 100% | 1 |
| 6 | ATERLIER BIOINFORMAT | 28324 | 0% | 100% | 1 |
| 7 | BASIC SCI BASIC SCI | 28324 | 0% | 100% | 1 |
| 8 | BIO ERS EVOLUCAO MOL | 28324 | 0% | 100% | 1 |
| 9 | BIOINFORMAT PHARMACOGENET | 28324 | 0% | 100% | 1 |
| 10 | BIOINFORMAT PHARMACOGENOMICS | 28324 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR BIOLOGY AND EVOLUTION | 276368 | 22% | 4% | 233 |
| 2 | JOURNAL OF MOLECULAR EVOLUTION | 51187 | 8% | 2% | 83 |
| 3 | BMC EVOLUTIONARY BIOLOGY | 22939 | 5% | 2% | 52 |
| 4 | GENOME BIOLOGY AND EVOLUTION | 17615 | 3% | 2% | 29 |
| 5 | EVOLUTIONARY BIOINFORMATICS | 12925 | 1% | 4% | 12 |
| 6 | BIOLOGY DIRECT | 8615 | 1% | 2% | 13 |
| 7 | SYSTEMATIC BIOLOGY | 5400 | 1% | 1% | 16 |
| 8 | BMC BIOINFORMATICS | 3350 | 3% | 0% | 30 |
| 9 | GENETICS | 2985 | 3% | 0% | 37 |
| 10 | PLOS COMPUTATIONAL BIOLOGY | 2667 | 2% | 0% | 21 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CODON MODELS | 768093 | 4% | 68% | 40 | Search CODON+MODELS | Search CODON+MODELS |
| 2 | POSITIVE SELECTION | 482331 | 14% | 11% | 152 | Search POSITIVE+SELECTION | Search POSITIVE+SELECTION |
| 3 | CODON SUBSTITUTION MODEL | 241362 | 1% | 61% | 14 | Search CODON+SUBSTITUTION+MODEL | Search CODON+SUBSTITUTION+MODEL |
| 4 | DN DS | 207683 | 2% | 33% | 22 | Search DN+DS | Search DN+DS |
| 5 | BRANCH SITE MODEL | 181273 | 1% | 80% | 8 | Search BRANCH+SITE+MODEL | Search BRANCH+SITE+MODEL |
| 6 | PROTEIN EVOLUTION | 166836 | 6% | 9% | 66 | Search PROTEIN+EVOLUTION | Search PROTEIN+EVOLUTION |
| 7 | SYNONYMOUS SUBSTITUTION | 139021 | 2% | 27% | 18 | Search SYNONYMOUS+SUBSTITUTION | Search SYNONYMOUS+SUBSTITUTION |
| 8 | BRANCH SITE TEST | 113297 | 0% | 100% | 4 | Search BRANCH+SITE+TEST | Search BRANCH+SITE+TEST |
| 9 | CLADE MODEL | 113297 | 0% | 100% | 4 | Search CLADE+MODEL | Search CLADE+MODEL |
| 10 | GENE COMPACTNESS | 113297 | 0% | 100% | 4 | Search GENE+COMPACTNESS | Search GENE+COMPACTNESS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ANISIMOVA, M , KOSIOL, C , (2009) INVESTIGATING PROTEIN-CODING SEQUENCE EVOLUTION WITH PROBABILISTIC CODON SUBSTITUTION MODELS.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 26. ISSUE 2. P. 255 -271 | 89 | 59% | 72 |
| 2 | YANG, ZH , (2007) PAML 4: PHYLOGENETIC ANALYSIS BY MAXIMUM LIKELIHOOD.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 24. ISSUE 8. P. 1586 -1591 | 27 | 48% | 3661 |
| 3 | ECHAVE, J , SPIELMAN, SJ , WILKE, CO , (2016) CAUSES OF EVOLUTIONARY RATE VARIATION AMONG PROTEIN SITES.NATURE REVIEWS GENETICS. VOL. 17. ISSUE 2. P. 109 -121 | 58 | 44% | 11 |
| 4 | DELPORT, W , SCHEFFLER, K , SEOIGHE, C , (2009) MODELS OF CODING SEQUENCE EVOLUTION.BRIEFINGS IN BIOINFORMATICS. VOL. 10. ISSUE 1. P. 97 -109 | 62 | 68% | 24 |
| 5 | MURRELL, B , WEAVER, S , SMITH, MD , WERTHEIM, JO , MURRELL, S , AYLWARD, A , EREN, K , POLLNER, T , MARTIN, DP , SMITH, DM , ET AL (2015) GENE-WIDE IDENTIFICATION OF EPISODIC SELECTION.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 32. ISSUE 5. P. 1365 -1371 | 25 | 96% | 18 |
| 6 | POND, SLK , MURRELL, B , FOURMENT, M , FROST, SDW , DELPORT, W , SCHEFFLER, K , (2011) A RANDOM EFFECTS BRANCH-SITE MODEL FOR DETECTING EPISODIC DIVERSIFYING SELECTION.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 28. ISSUE 11. P. 3033 -3043 | 28 | 82% | 107 |
| 7 | SPIELMAN, SJ , WAN, SY , WILKE, CO , (2016) A COMPARISON OF ONE-RATE AND TWO-RATE INFERENCE FRAMEWORKS FOR SITE-SPECIFIC DN/DS ESTIMATION.GENETICS. VOL. 204. ISSUE 2. P. 499 -+ | 30 | 79% | 2 |
| 8 | ANISIMOVA, M , LIBERLES, DA , (2007) THE QUEST FOR NATURAL SELECTION IN THE AGE OF COMPARATIVE GENOMICS.HEREDITY. VOL. 99. ISSUE 6. P. 567 -579 | 68 | 45% | 46 |
| 9 | BLOOM, JD , (2017) IDENTIFICATION OF POSITIVE SELECTION IN GENES IS GREATLY IMPROVED BY USING EXPERIMENTALLY INFORMED SITE-SPECIFIC MODELS.BIOLOGY DIRECT. VOL. 12. ISSUE . P. - | 48 | 55% | 0 |
| 10 | ALVAREZ-PONCE, D , SABATER-MUNOZ, B , TOFT, C , RUIZ-GONZALEZ, MX , FARES, MA , (2016) ESSENTIALITY IS A STRONG DETERMINANT OF PROTEIN RATES OF EVOLUTION DURING MUTATION ACCUMULATION EXPERIMENTS IN ESCHERICHIA COLI.GENOME BIOLOGY AND EVOLUTION. VOL. 8. ISSUE 9. P. 2914 -2927 | 50 | 52% | 0 |
Classes with closest relation at Level 1 |