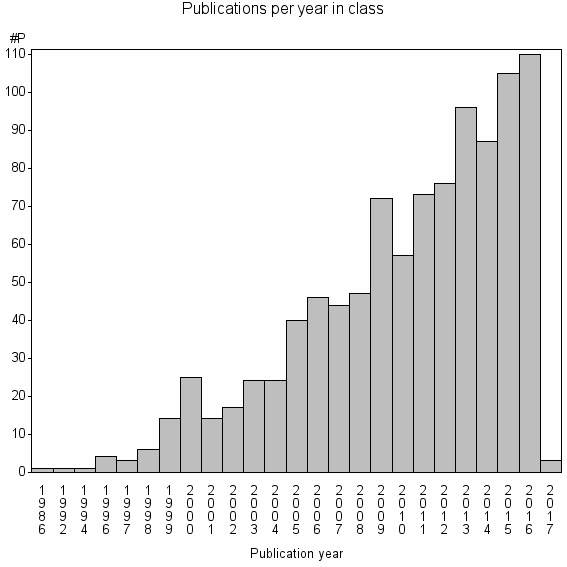
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 11331 | 990 | 43.5 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 11331 | 1 | NSSNPS//POLYPHEN//LSDB | 990 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | NSSNPS | authKW | 510979 | 3% | 61% | 27 |
| 2 | POLYPHEN | authKW | 260608 | 1% | 65% | 13 |
| 3 | LSDB | authKW | 257006 | 2% | 56% | 15 |
| 4 | HUMAN MUTATION | journal | 150327 | 14% | 4% | 139 |
| 5 | PATHOGENICITY PREDICTION | authKW | 146946 | 1% | 53% | 9 |
| 6 | SNYDER ROBINSON SYNDROME | authKW | 138787 | 1% | 75% | 6 |
| 7 | MUTATION DATABASE | authKW | 133011 | 2% | 25% | 17 |
| 8 | BRISTOL SYST BIOMED | address | 123369 | 0% | 100% | 4 |
| 9 | POLYPHEN 2 | authKW | 110148 | 1% | 71% | 5 |
| 10 | DBNSFP | authKW | 92527 | 0% | 100% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 14781 | 23% | 0% | 223 |
| 2 | Genetics & Heredity | 7212 | 36% | 0% | 354 |
| 3 | Biochemical Research Methods | 3714 | 21% | 0% | 207 |
| 4 | Biotechnology & Applied Microbiology | 3439 | 26% | 0% | 254 |
| 5 | Biochemistry & Molecular Biology | 1577 | 35% | 0% | 348 |
| 6 | Biophysics | 247 | 7% | 0% | 67 |
| 7 | Medical Informatics | 61 | 1% | 0% | 11 |
| 8 | Computer Science, Interdisciplinary Applications | 25 | 2% | 0% | 20 |
| 9 | Cell Biology | 14 | 4% | 0% | 41 |
| 10 | Medicine, Research & Experimental | 4 | 2% | 0% | 20 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BRISTOL SYST BIOMED | 123369 | 0% | 100% | 4 |
| 2 | NETWORK BIOMED RARE DIS CIBERER | 92527 | 0% | 100% | 3 |
| 3 | BIOTECHNOL CHEM BIOMED ENGN | 80578 | 1% | 19% | 14 |
| 4 | GENOM DISORDERS | 55963 | 1% | 26% | 7 |
| 5 | HUMAN VARIOME PROJECT | 55514 | 0% | 60% | 3 |
| 6 | CELL BIOL GENETMGC | 41122 | 0% | 67% | 2 |
| 7 | PL INFORMAT MANAGEMENT ECON | 41122 | 0% | 67% | 2 |
| 8 | BIO SCI TECHNOL | 35848 | 2% | 6% | 21 |
| 9 | AFFILIATED HOSP 1BEIJING GENOME CAS | 30842 | 0% | 100% | 1 |
| 10 | AGR BYPROD PROC | 30842 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | HUMAN MUTATION | 150327 | 14% | 4% | 139 |
| 2 | BIOINFORMATICS | 24457 | 9% | 1% | 92 |
| 3 | INTERDISCIPLINARY SCIENCES-COMPUTATIONAL LIFE SCIENCES | 19537 | 1% | 5% | 14 |
| 4 | BMC BIOINFORMATICS | 11052 | 5% | 1% | 52 |
| 5 | NUCLEIC ACIDS RESEARCH | 6104 | 9% | 0% | 89 |
| 6 | BMC GENOMICS | 3255 | 3% | 0% | 32 |
| 7 | DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION | 2308 | 1% | 1% | 7 |
| 8 | GENOME MEDICINE | 2297 | 1% | 1% | 7 |
| 9 | HUMAN GENOMICS | 2163 | 0% | 2% | 3 |
| 10 | BRIEFINGS IN BIOINFORMATICS | 2161 | 1% | 1% | 7 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NSSNPS | 510979 | 3% | 61% | 27 | Search NSSNPS | Search NSSNPS |
| 2 | POLYPHEN | 260608 | 1% | 65% | 13 | Search POLYPHEN | Search POLYPHEN |
| 3 | LSDB | 257006 | 2% | 56% | 15 | Search LSDB | Search LSDB |
| 4 | PATHOGENICITY PREDICTION | 146946 | 1% | 53% | 9 | Search PATHOGENICITY+PREDICTION | Search PATHOGENICITY+PREDICTION |
| 5 | SNYDER ROBINSON SYNDROME | 138787 | 1% | 75% | 6 | Search SNYDER+ROBINSON+SYNDROME | Search SNYDER+ROBINSON+SYNDROME |
| 6 | MUTATION DATABASE | 133011 | 2% | 25% | 17 | Search MUTATION+DATABASE | Search MUTATION+DATABASE |
| 7 | POLYPHEN 2 | 110148 | 1% | 71% | 5 | Search POLYPHEN+2 | Search POLYPHEN+2 |
| 8 | DBNSFP | 92527 | 0% | 100% | 3 | Search DBNSFP | Search DBNSFP |
| 9 | FATHMM | 92527 | 0% | 100% | 3 | Search FATHMM | Search FATHMM |
| 10 | SNPSGO | 92527 | 0% | 100% | 3 | Search SNPSGO | Search SNPSGO |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | NIROULA, A , VIHINEN, M , (2016) VARIATION INTERPRETATION PREDICTORS: PRINCIPLES, TYPES, PERFORMANCE, AND CHOICE.HUMAN MUTATION. VOL. 37. ISSUE 6. P. 579 -597 | 111 | 46% | 6 |
| 2 | KUMAR, A , RAJENDRAN, V , SETHUMADHAVAN, R , SHUKLA, P , TIWARI, S , PUROHIT, R , (2014) COMPUTATIONAL SNP ANALYSIS: CURRENT APPROACHES AND FUTURE PROSPECTS.CELL BIOCHEMISTRY AND BIOPHYSICS. VOL. 68. ISSUE 2. P. 233-239 | 43 | 88% | 6 |
| 3 | RIERA, C , LOIS, S , DE LA CRUZ, X , (2014) PREDICTION OF PATHOLOGICAL MUTATIONS IN PROTEINS: THE CHALLENGE OF INTEGRATING SEQUENCE CONSERVATION AND STRUCTURE STABILITY PRINCIPLES.WILEY INTERDISCIPLINARY REVIEWS-COMPUTATIONAL MOLECULAR SCIENCE. VOL. 4. ISSUE 3. P. 249 -268 | 64 | 52% | 2 |
| 4 | KULSHRESHTHA, S , CHAUDHARY, V , GOSWAMI, GK , MATHUR, N , (2016) COMPUTATIONAL APPROACHES FOR PREDICTING MUTANT PROTEIN STABILITY.JOURNAL OF COMPUTER-AIDED MOLECULAR DESIGN. VOL. 30. ISSUE 5. P. 401 -412 | 45 | 71% | 0 |
| 5 | WANG, MJ , SUN, ZW , AKUTSU, T , SONG, JM , (2013) RECENT ADVANCES IN PREDICTING FUNCTIONAL IMPACT OF SINGLE AMINO ACID POLYMORPHISMS: A REVIEW OF USEFUL FEATURES, COMPUTATIONAL METHODS AND AVAILABLE TOOLS.CURRENT BIOINFORMATICS. VOL. 8. ISSUE 2. P. 161 -176 | 56 | 59% | 6 |
| 6 | TANG, HM , THOMAS, PD , (2016) TOOLS FOR PREDICTING THE FUNCTIONAL IMPACT OF NONSYNONYMOUS GENETIC VARIATION.GENETICS. VOL. 203. ISSUE 2. P. 635 -647 | 55 | 52% | 0 |
| 7 | PONS, T , VAZQUEZ, M , MATEY-HERNANDEZ, ML , BRUNAK, S , VALENCIA, A , IZARZUGAZA, JMG , (2016) KINMUTRF: A RANDOM FOREST CLASSIFIER OF SEQUENCE VARIANTS IN THE HUMAN PROTEIN KINASE SUPERFAMILY.BMC GENOMICS. VOL. 17. ISSUE . P. - | 40 | 70% | 0 |
| 8 | PETERSON, TA , DOUGHTY, E , KANN, MG , (2013) TOWARDS PRECISION MEDICINE: ADVANCES IN COMPUTATIONAL APPROACHES FOR THE ANALYSIS OF HUMAN VARIANTS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 425. ISSUE 21. P. 4047 -4063 | 60 | 40% | 31 |
| 9 | CASTELLANA, S , MAZZA, T , (2013) CONGRUENCY IN THE PREDICTION OF PATHOGENIC MISSENSE MUTATIONS: STATE-OF-THE-ART WEB-BASED TOOLS.BRIEFINGS IN BIOINFORMATICS. VOL. 14. ISSUE 4. P. 448 -459 | 34 | 72% | 27 |
| 10 | KUMAR, P , HENIKOFF, S , NG, PC , (2009) PREDICTING THE EFFECTS OF CODING NON-SYNONYMOUS VARIANTS ON PROTEIN FUNCTION USING THE SIFT ALGORITHM.NATURE PROTOCOLS. VOL. 4. ISSUE 7. P. 1073-1082 | 12 | 71% | 2135 |
Classes with closest relation at Level 1 |