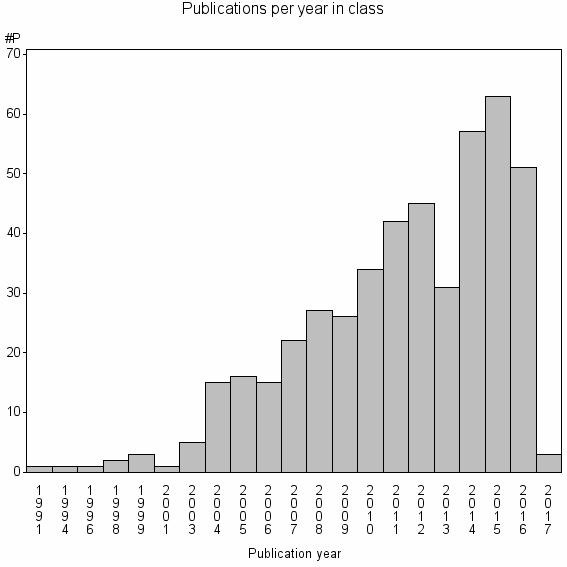
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 20432 | 461 | 48.1 | 89% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 219 | 3 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN FOLDING//PROTEIN STRUCTURE PREDICTION | 50409 |
| 365 | 2 | PROTEIN STRUCTURE PREDICTION//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PSEUDO AMINO ACID COMPOSITION | 17331 |
| 20432 | 1 | KINASE SPECIFIC//PTM CODE//CELLULAR MOL LOG TEAM | 461 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | KINASE SPECIFIC | authKW | 198709 | 1% | 100% | 3 |
| 2 | PTM CODE | authKW | 198709 | 1% | 100% | 3 |
| 3 | CELLULAR MOL LOG TEAM | address | 150531 | 1% | 45% | 5 |
| 4 | PHOSPHORYLATION NETWORK | authKW | 149030 | 1% | 75% | 3 |
| 5 | PHOSPHORYLATION PREDICTION | authKW | 132472 | 0% | 100% | 2 |
| 6 | POSITION WEIGHT AMINO ACID COMPOSITION | authKW | 132472 | 0% | 100% | 2 |
| 7 | POST TRANSLATIONAL MODIFICATIONS OF PROTEINS PTMS | authKW | 132472 | 0% | 100% | 2 |
| 8 | PROFILE PROFILE SEQUENCE SIMILARITY PSI BLAST FFAS | authKW | 132472 | 0% | 100% | 2 |
| 9 | SOL H SNYDER NEUROSCI | address | 132472 | 0% | 100% | 2 |
| 10 | SOLVENT ACCESSIBLE SURFACE AREA ASA | authKW | 132472 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 10517 | 28% | 0% | 128 |
| 2 | Biochemical Research Methods | 5096 | 35% | 0% | 162 |
| 3 | Biotechnology & Applied Microbiology | 1425 | 24% | 0% | 112 |
| 4 | Biochemistry & Molecular Biology | 1384 | 46% | 0% | 213 |
| 5 | Cell Biology | 215 | 13% | 0% | 59 |
| 6 | Genetics & Heredity | 119 | 8% | 0% | 37 |
| 7 | Biology | 86 | 5% | 0% | 21 |
| 8 | Biophysics | 66 | 5% | 0% | 25 |
| 9 | Computer Science, Interdisciplinary Applications | 30 | 3% | 0% | 13 |
| 10 | Evolutionary Biology | 19 | 2% | 0% | 8 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CELLULAR MOL LOG TEAM | 150531 | 1% | 45% | 5 |
| 2 | SOL H SNYDER NEUROSCI | 132472 | 0% | 100% | 2 |
| 3 | MOL BIOINFORMAT | 73578 | 2% | 11% | 10 |
| 4 | ADV CHEM PHYS BIOL PHARMACEUT SCI | 66236 | 0% | 100% | 1 |
| 5 | BIOPHYSEPIGEN TRANSLAT PROGRAM | 66236 | 0% | 100% | 1 |
| 6 | CHROMATOG RANDA | 66236 | 0% | 100% | 1 |
| 7 | COMP INTELLIGENCE LEARNING DISCOVERY | 66236 | 0% | 100% | 1 |
| 8 | CYBERNET INTELLIGENCE | 66236 | 0% | 100% | 1 |
| 9 | EPIGENET CHROMATIN DYNAMEPIGEN TRANSLAT PR | 66236 | 0% | 100% | 1 |
| 10 | GENOME ORIENTED BIOINFORRNAT | 66236 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 12016 | 10% | 0% | 44 |
| 2 | BMC BIOINFORMATICS | 11411 | 8% | 0% | 36 |
| 3 | DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION | 6496 | 2% | 1% | 8 |
| 4 | SCIENCE SIGNALING | 5176 | 2% | 1% | 11 |
| 5 | MOLECULAR & CELLULAR PROTEOMICS | 4817 | 3% | 0% | 15 |
| 6 | MOLECULAR BIOSYSTEMS | 2435 | 2% | 0% | 10 |
| 7 | BRIEFINGS IN BIOINFORMATICS | 2373 | 1% | 1% | 5 |
| 8 | NUCLEIC ACIDS RESEARCH | 2258 | 8% | 0% | 37 |
| 9 | MOLECULAR SYSTEMS BIOLOGY | 2180 | 1% | 1% | 5 |
| 10 | PLOS COMPUTATIONAL BIOLOGY | 1417 | 2% | 0% | 10 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | TROST, B , KUSALIK, A , (2011) COMPUTATIONAL PREDICTION OF EUKARYOTIC PHOSPHORYLATION SITES.BIOINFORMATICS. VOL. 27. ISSUE 21. P. 2927 -2935 | 60 | 63% | 52 |
| 2 | HUANG, GH , LI, XM , (2015) A REVIEW OF COMPUTATIONAL IDENTIFICATION OF PROTEIN POST-TRANSLATIONAL MODIFICATIONS.MINI-REVIEWS IN ORGANIC CHEMISTRY. VOL. 12. ISSUE 6. P. 468 -480 | 77 | 51% | 0 |
| 3 | PALMERI, A , FERRE, F , HELMER-CITTERICH, M , (2014) EXPLOITING HOLISTIC APPROACHES TO MODEL SPECIFICITY IN PROTEIN PHOSPHORYLATION.FRONTIERS IN GENETICS. VOL. 5. ISSUE . P. - | 51 | 57% | 2 |
| 4 | XUE, Y , GAO, XJ , CAO, J , LIU, ZX , JIN, CJ , WEN, LP , YAO, XB , REN, JA , (2010) A SUMMARY OF COMPUTATIONAL RESOURCES FOR PROTEIN PHOSPHORYLATION.CURRENT PROTEIN & PEPTIDE SCIENCE. VOL. 11. ISSUE 6. P. 485-496 | 51 | 53% | 23 |
| 5 | GONG, HP , LIU, XQ , WU, J , HE, ZY , (2014) DATA CONSTRUCTION FOR PHOSPHORYLATION SITE PREDICTION.BRIEFINGS IN BIOINFORMATICS. VOL. 15. ISSUE 5. P. 839 -855 | 27 | 87% | 0 |
| 6 | SU, MG , LEE, TY , (2013) INCORPORATING SUBSTRATE SEQUENCE MOTIFS AND SPATIAL AMINO ACID COMPOSITION TO IDENTIFY KINASE-SPECIFIC PHOSPHORYLATION SITES ON PROTEIN THREE-DIMENSIONAL STRUCTURES.BMC BIOINFORMATICS. VOL. 14. ISSUE . P. - | 30 | 75% | 5 |
| 7 | HUANG, KY , SU, MG , KAO, HJ , HSIEH, YC , JHONG, JH , CHENG, KH , HUANG, HD , LEE, TY , (2016) DBPTM 2016: 10-YEAR ANNIVERSARY OF A RESOURCE FOR POST-TRANSLATIONAL MODIFICATION OF PROTEINS.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D435 -D446 | 31 | 48% | 6 |
| 8 | VESELOVSKY, AV , POROIKOV, VV , SOBOLEV, BN , (2014) PREDICTION OF PROTEIN POST-TRANSLATIONAL MODIFICATIONS: MAIN TRENDS AND METHODS.RUSSIAN CHEMICAL REVIEWS. VOL. 83. ISSUE 2. P. 143 -154 | 50 | 41% | 2 |
| 9 | TROST, B , MALEKI, F , KUSALIK, A , NAPPER, S , (2016) DAPPLE 2: A TOOL FOR THE HOMOLOGY-BASED PREDICTION OF POST-TRANSLATIONAL MODIFICATION SITES.JOURNAL OF PROTEOME RESEARCH. VOL. 15. ISSUE 8. P. 2760 -2767 | 24 | 69% | 1 |
| 10 | WANG, MH , JIANG, YJ , XU, XY , (2015) A NOVEL METHOD FOR PREDICTING POST-TRANSLATIONAL MODIFICATIONS ON SERINE AND THREONINE SITES BY USING SITE-MODIFICATION NETWORK PROFILES.MOLECULAR BIOSYSTEMS. VOL. 11. ISSUE 11. P. 3092 -3100 | 24 | 63% | 2 |
Classes with closest relation at Level 1 |