Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
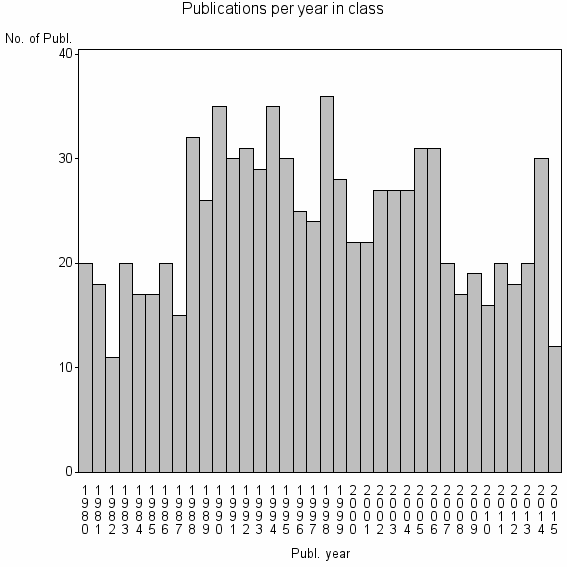
| 12225 | 858 | 42.9 | 75% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | FRAMESHIFTING | Author keyword | 66 | 63% | 8% | 66 |
| 2 | RIBOSOMAL FRAMESHIFTING | Author keyword | 21 | 61% | 3% | 23 |
| 3 | RNA PSEUDOKNOT | Author keyword | 18 | 55% | 3% | 23 |
| 4 | RECODING | Author keyword | 11 | 33% | 3% | 28 |
| 5 | 1 RIBOSOMAL FRAMESHIFTING | Author keyword | 8 | 100% | 1% | 5 |
| 6 | GENET MOL TRADUCT | Address | 6 | 80% | 0% | 4 |
| 7 | SLIPPERY SEQUENCE | Author keyword | 6 | 100% | 0% | 4 |
| 8 | PSEUDOKNOT | Author keyword | 5 | 16% | 3% | 27 |
| 9 | 1 FRAMESHIFT | Author keyword | 4 | 75% | 0% | 3 |
| 10 | HUNGRY CODON | Author keyword | 4 | 75% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | FRAMESHIFTING | 66 | 63% | 8% | 66 | Search FRAMESHIFTING | Search FRAMESHIFTING |
| 2 | RIBOSOMAL FRAMESHIFTING | 21 | 61% | 3% | 23 | Search RIBOSOMAL+FRAMESHIFTING | Search RIBOSOMAL+FRAMESHIFTING |
| 3 | RNA PSEUDOKNOT | 18 | 55% | 3% | 23 | Search RNA+PSEUDOKNOT | Search RNA+PSEUDOKNOT |
| 4 | RECODING | 11 | 33% | 3% | 28 | Search RECODING | Search RECODING |
| 5 | 1 RIBOSOMAL FRAMESHIFTING | 8 | 100% | 1% | 5 | Search 1+RIBOSOMAL+FRAMESHIFTING | Search 1+RIBOSOMAL+FRAMESHIFTING |
| 6 | SLIPPERY SEQUENCE | 6 | 100% | 0% | 4 | Search SLIPPERY+SEQUENCE | Search SLIPPERY+SEQUENCE |
| 7 | PSEUDOKNOT | 5 | 16% | 3% | 27 | Search PSEUDOKNOT | Search PSEUDOKNOT |
| 8 | 1 FRAMESHIFT | 4 | 75% | 0% | 3 | Search 1+FRAMESHIFT | Search 1+FRAMESHIFT |
| 9 | HUNGRY CODON | 4 | 75% | 0% | 3 | Search HUNGRY+CODON | Search HUNGRY+CODON |
| 10 | PROGRAMMED FRAMESHIFTING | 3 | 57% | 0% | 4 | Search PROGRAMMED+FRAMESHIFTING | Search PROGRAMMED+FRAMESHIFTING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RELEASE FACTOR II | 64 | 65% | 7% | 61 |
| 2 | STIMULATORY SIGNAL | 51 | 91% | 2% | 21 |
| 3 | SIMIAN RETROVIRUS 1 | 31 | 92% | 1% | 12 |
| 4 | RNA PSEUDOKNOT | 26 | 56% | 4% | 32 |
| 5 | UGA SUPPRESSION | 26 | 80% | 2% | 16 |
| 6 | PROGRAMMED 1 RIBOSOMAL FRAMESHIFT | 21 | 73% | 2% | 16 |
| 7 | RIBOSOMAL FRAMESHIFT | 21 | 65% | 2% | 20 |
| 8 | CODING GAP | 18 | 89% | 1% | 8 |
| 9 | GENE AMBER CODON | 15 | 88% | 1% | 7 |
| 10 | TRANSLATIONAL READTHROUGH | 13 | 58% | 2% | 15 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Frameshifting RNA pseudoknots: Structure and mechanism | 2009 | 101 | 146 | 48% |
| Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting | 2000 | 141 | 103 | 62% |
| Recoding in bacteriophages and bacterial IS elements | 2006 | 50 | 37 | 68% |
| Reprogrammed genetic decoding in cellular gene expression | 2004 | 152 | 71 | 41% |
| Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV | 2006 | 59 | 137 | 50% |
| Mechanisms and implications of programmed translational frameshifting | 2012 | 25 | 86 | 65% |
| How translational accuracy influences reading frame maintenance | 1999 | 78 | 42 | 64% |
| Recoding: translational bifurcations in gene expression | 2002 | 121 | 127 | 42% |
| Targeting frameshifting in the human immunodeficiency virus | 2012 | 10 | 60 | 73% |
| Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs | 2001 | 113 | 96 | 52% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENET MOL TRADUCT | 6 | 80% | 0.5% | 4 |
| 2 | PROGRAM MOL BIOSCI | 1 | 16% | 0.7% | 6 |
| 3 | MACROMOL DESIGN | 1 | 15% | 0.7% | 6 |
| 4 | HEPATITIS RELATED EMERGING AGENTS | 1 | 23% | 0.3% | 3 |
| 5 | OFF BLOOD REVIEWCBER | 1 | 50% | 0.1% | 1 |
| 6 | PROGRAM COMPUTAT MOL BIOL STUDIES | 1 | 50% | 0.1% | 1 |
| 7 | PROGRAMS MOL BIOSCI | 1 | 50% | 0.1% | 1 |
| 8 | SECT GENEXP S | 1 | 50% | 0.1% | 1 |
| 9 | STRUCT BIOL GEN | 1 | 50% | 0.1% | 1 |
| 10 | UMR5100 | 1 | 10% | 0.7% | 6 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000182142 | SUP35//TRANSLATION TERMINATION//URE3 |
| 2 | 0.0000135595 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 3 | 0.0000128356 | TRANS TRANSLATION//TMRNA//SMPB |
| 4 | 0.0000127789 | QUEUINE//TRNA MODIFICATION//RNA MODIFICATION |
| 5 | 0.0000112667 | TY1//SECT EUKARYOT TRANSPOSABLE ELEMENTS//TY ELEMENTS |
| 6 | 0.0000102711 | TRANSLATIONAL SELECTION//CODON USAGE//SYNONYMOUS CODON USAGE |
| 7 | 0.0000093504 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 8 | 0.0000092753 | PINEAL GLAND PEPTIDES//SUBSTRAIN DIFFERENCE//CAROLINA ENVIRONM BIOINFORMAT |
| 9 | 0.0000076519 | RNA SECONDARY STRUCTURE//RNA SECONDARY STRUCTURE PREDICTION//RNA STRUCTURE PREDICTION |
| 10 | 0.0000072648 | SD SEQUENCE//RIBOSOMAL PROTEIN S1//SHINE DALGARNO SEQUENCE |