Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
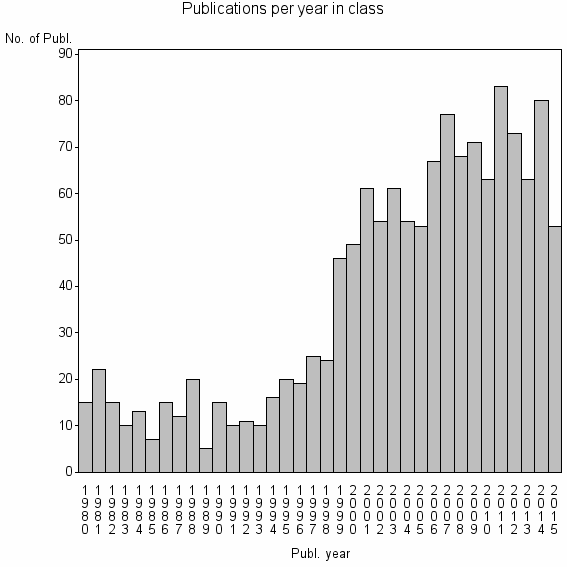
| 6844 | 1360 | 48.6 | 81% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SUP35 | Author keyword | 131 | 87% | 5% | 65 |
| 2 | TRANSLATION TERMINATION | Author keyword | 72 | 53% | 7% | 95 |
| 3 | URE3 | Author keyword | 72 | 93% | 2% | 27 |
| 4 | YEAST PRION | Author keyword | 58 | 80% | 3% | 36 |
| 5 | SUP45 | Author keyword | 57 | 95% | 1% | 19 |
| 6 | ERF1 | Author keyword | 50 | 75% | 3% | 36 |
| 7 | URE2P | Author keyword | 44 | 76% | 2% | 31 |
| 8 | SUP35P | Author keyword | 32 | 85% | 1% | 17 |
| 9 | STOP CODON RECOGNITION | Author keyword | 32 | 88% | 1% | 15 |
| 10 | ERF3 | Author keyword | 28 | 64% | 2% | 28 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SUP35 | 131 | 87% | 5% | 65 | Search SUP35 | Search SUP35 |
| 2 | TRANSLATION TERMINATION | 72 | 53% | 7% | 95 | Search TRANSLATION+TERMINATION | Search TRANSLATION+TERMINATION |
| 3 | URE3 | 72 | 93% | 2% | 27 | Search URE3 | Search URE3 |
| 4 | YEAST PRION | 58 | 80% | 3% | 36 | Search YEAST+PRION | Search YEAST+PRION |
| 5 | SUP45 | 57 | 95% | 1% | 19 | Search SUP45 | Search SUP45 |
| 6 | ERF1 | 50 | 75% | 3% | 36 | Search ERF1 | Search ERF1 |
| 7 | URE2P | 44 | 76% | 2% | 31 | Search URE2P | Search URE2P |
| 8 | SUP35P | 32 | 85% | 1% | 17 | Search SUP35P | Search SUP35P |
| 9 | STOP CODON RECOGNITION | 32 | 88% | 1% | 15 | Search STOP+CODON+RECOGNITION | Search STOP+CODON+RECOGNITION |
| 10 | ERF3 | 28 | 64% | 2% | 28 | Search ERF3 | Search ERF3 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PSI PRION | 638 | 95% | 16% | 212 |
| 2 | DE NOVO APPEARANCE | 385 | 97% | 8% | 107 |
| 3 | SUP35 GENE | 269 | 97% | 5% | 74 |
| 4 | SUP35 PROTEIN | 263 | 92% | 8% | 104 |
| 5 | TRANSFER RNA HYDROLYSIS | 103 | 78% | 5% | 69 |
| 6 | RELEASE FACTOR ERF1 | 102 | 76% | 5% | 71 |
| 7 | URE3 PRION | 94 | 97% | 2% | 28 |
| 8 | YEAST PSI PRION | 89 | 90% | 3% | 38 |
| 9 | CHAIN RELEASE FACTORS | 87 | 81% | 4% | 52 |
| 10 | SUP35 | 82 | 77% | 4% | 55 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PRION | 13 | 17% | 5% | 67 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Yeast Prions: Structure, Biology, and Prion-Handling Systems | 2015 | 1 | 199 | 75% |
| Prions of fungi: inherited structures and biological roles | 2007 | 128 | 96 | 70% |
| Prions, protein homeostasis, and phenotypic diversity | 2010 | 62 | 76 | 71% |
| Newly identified prions in budding yeast, and their possible functions | 2011 | 25 | 75 | 73% |
| Blessings in disguise: biological benefits of prion-like mechanisms | 2013 | 22 | 87 | 49% |
| The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility | 2011 | 29 | 67 | 64% |
| Prions as adaptive conduits of memory and inheritance | 2005 | 238 | 143 | 47% |
| Termination of translation: interplay of mRNA, rRNAs and release factors? | 2003 | 150 | 67 | 70% |
| Analysis of amyloid aggregates using agarose gel electrophoresis | 2006 | 55 | 47 | 77% |
| Modulation and elimination of yeast prions by protein chaperones and co-chaperones | 2011 | 14 | 55 | 82% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENET MOL CHAMPIGNONS | 12 | 50% | 1.3% | 18 |
| 2 | AMYLOID BIOL | 6 | 80% | 0.3% | 4 |
| 3 | KENT FUNGAL GRP | 5 | 42% | 0.7% | 10 |
| 4 | CNRSUMR 5095 | 5 | 55% | 0.4% | 6 |
| 5 | PROT CONFORMAT DIS | 4 | 67% | 0.3% | 4 |
| 6 | BIOCHIM GENET CELLULAIRE | 4 | 34% | 0.7% | 10 |
| 7 | BIOENGN BIOSCI | 3 | 11% | 2.0% | 27 |
| 8 | TANAKA UNIT | 3 | 60% | 0.2% | 3 |
| 9 | IBGCUMR 5095 | 2 | 67% | 0.1% | 2 |
| 10 | MOL BIOLMINATO KU | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000182142 | FRAMESHIFTING//RIBOSOMAL FRAMESHIFTING//RNA PSEUDOKNOT |
| 2 | 0.0000138040 | CELLULAR NEUROL//PROTEOPATHY//PRION LIKE AGGREGATION |
| 3 | 0.0000089871 | TRANS TRANSLATION//TMRNA//SMPB |
| 4 | 0.0000077715 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 5 | 0.0000070669 | AMYLOID FIBRIL//AMYLOID FIBRILS//AMYLOID |
| 6 | 0.0000068596 | ATP DEPENDENT PROTEASES//CLPB//FTSH |
| 7 | 0.0000063673 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 8 | 0.0000063062 | DNAJ//GRPE//DNAK |
| 9 | 0.0000062731 | PRION PROTEIN//PRION//CELLULAR PRION PROTEIN |
| 10 | 0.0000053738 | EEF1A2//ELONGATION FACTOR 1 GAMMA//ELONGATION FACTOR 1 |