Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
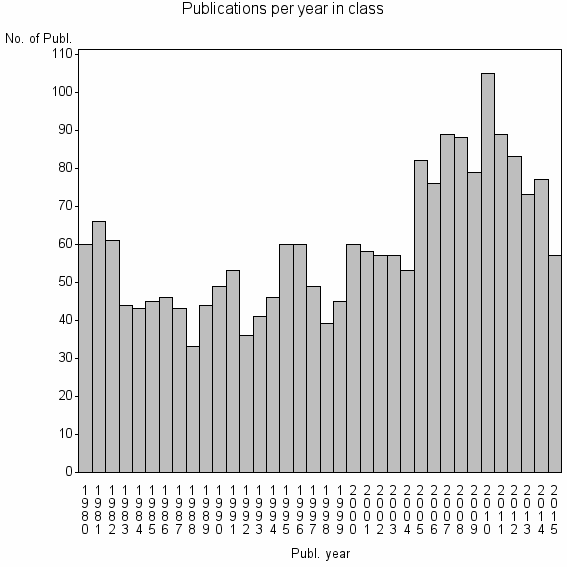
| 2527 | 2146 | 42.1 | 71% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | ELONGATION FACTOR TU | Author keyword | 76 | 65% | 3% | 73 |
| 2 | EF G | Author keyword | 47 | 79% | 1% | 30 |
| 3 | ELONGATION FACTOR G | Author keyword | 38 | 70% | 1% | 32 |
| 4 | RIBOSOME | Author keyword | 29 | 14% | 9% | 199 |
| 5 | RIBOSOME RECYCLING FACTOR | Author keyword | 23 | 69% | 1% | 20 |
| 6 | EF TU | Author keyword | 23 | 47% | 2% | 36 |
| 7 | IF2 | Author keyword | 19 | 70% | 1% | 16 |
| 8 | ELONGATION FACTOR TS | Author keyword | 19 | 76% | 1% | 13 |
| 9 | HYBRID STATE | Author keyword | 19 | 76% | 1% | 13 |
| 10 | KIRROMYCIN | Author keyword | 19 | 64% | 1% | 18 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ELONGATION FACTOR TU | 76 | 65% | 3% | 73 | Search ELONGATION+FACTOR+TU | Search ELONGATION+FACTOR+TU |
| 2 | EF G | 47 | 79% | 1% | 30 | Search EF+G | Search EF+G |
| 3 | ELONGATION FACTOR G | 38 | 70% | 1% | 32 | Search ELONGATION+FACTOR+G | Search ELONGATION+FACTOR+G |
| 4 | RIBOSOME | 29 | 14% | 9% | 199 | Search RIBOSOME | Search RIBOSOME |
| 5 | RIBOSOME RECYCLING FACTOR | 23 | 69% | 1% | 20 | Search RIBOSOME+RECYCLING+FACTOR | Search RIBOSOME+RECYCLING+FACTOR |
| 6 | EF TU | 23 | 47% | 2% | 36 | Search EF+TU | Search EF+TU |
| 7 | IF2 | 19 | 70% | 1% | 16 | Search IF2 | Search IF2 |
| 8 | ELONGATION FACTOR TS | 19 | 76% | 1% | 13 | Search ELONGATION+FACTOR+TS | Search ELONGATION+FACTOR+TS |
| 9 | HYBRID STATE | 19 | 76% | 1% | 13 | Search HYBRID+STATE | Search HYBRID+STATE |
| 10 | KIRROMYCIN | 19 | 64% | 1% | 18 | Search KIRROMYCIN | Search KIRROMYCIN |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AMINOACYL TRANSFER RNA | 230 | 46% | 17% | 371 |
| 2 | ELONGATION FACTOR G | 139 | 48% | 10% | 216 |
| 3 | 70S RIBOSOME | 135 | 53% | 8% | 180 |
| 4 | HYBRID STATE | 120 | 83% | 3% | 68 |
| 5 | FACTOR EF TU | 100 | 55% | 6% | 125 |
| 6 | EF TU | 88 | 43% | 7% | 155 |
| 7 | EF G | 88 | 58% | 5% | 102 |
| 8 | KIRROMYCIN | 80 | 80% | 2% | 49 |
| 9 | ELONGATION FACTOR TU | 66 | 24% | 11% | 241 |
| 10 | MESSENGER RNA TRANSLOCATION | 63 | 83% | 2% | 35 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| What recent ribosome structures have revealed about the mechanism of translation | 2009 | 254 | 126 | 80% |
| Ribosome-targeting antibiotics and mechanisms of bacterial resistance | 2014 | 40 | 135 | 36% |
| Structural Basis of the Translational Elongation Cycle | 2013 | 33 | 174 | 76% |
| A structural understanding of the dynamic ribosome machine | 2008 | 153 | 48 | 65% |
| Ribosome structure and the mechanism of translation | 2002 | 402 | 96 | 51% |
| EF-G and EF4: translocation and back-translocation on the bacterial ribosome | 2014 | 11 | 103 | 69% |
| How Should We Think About the Ribosome? | 2012 | 24 | 64 | 80% |
| The ribosomal peptidyl transferase | 2007 | 73 | 54 | 78% |
| Structure and Dynamics of a Processive Brownian Motor: The Translating Ribosome | 2010 | 60 | 159 | 59% |
| Evolutionary optimization of speed and accuracy of decoding on the ribosome | 2011 | 29 | 67 | 69% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOL MCA | 9 | 31% | 1.1% | 23 |
| 2 | PHYS BIOCHEM | 8 | 11% | 3.4% | 74 |
| 3 | MOL BIOL RNA | 6 | 17% | 1.5% | 33 |
| 4 | HLTH INC | 6 | 40% | 0.6% | 12 |
| 5 | ABT VINGRON | 5 | 60% | 0.3% | 6 |
| 6 | GENOM RNOM | 4 | 31% | 0.6% | 12 |
| 7 | RIBOSOMAL ORIGINS EVOLUT | 3 | 57% | 0.2% | 4 |
| 8 | BIOCHEM BIOCHEM WASSENAARSEWEG 64 | 3 | 100% | 0.1% | 3 |
| 9 | ULTRASTRUKTURNETZWERK | 3 | 32% | 0.4% | 8 |
| 10 | INNSBRUCK BIO | 3 | 20% | 0.6% | 12 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000220232 | MRNA ANALOG//80S RIBOSOME//HUMAN RIBOSOME |
| 2 | 0.0000135595 | FRAMESHIFTING//RIBOSOMAL FRAMESHIFTING//RNA PSEUDOKNOT |
| 3 | 0.0000126966 | TRANS TRANSLATION//TMRNA//SMPB |
| 4 | 0.0000124511 | DAP3//ICT1//MITOCHONDRIAL RIBOSOMAL PROTEINS |
| 5 | 0.0000117640 | RIBOSOMAL STALK//ACIDIC RIBOSOMAL PROTEIN//ACIDIC RIBOSOMAL P PROTEINS |
| 6 | 0.0000113758 | SD SEQUENCE//RIBOSOMAL PROTEIN S1//SHINE DALGARNO SEQUENCE |
| 7 | 0.0000111407 | EEF1A2//ELONGATION FACTOR 1 GAMMA//ELONGATION FACTOR 1 |
| 8 | 0.0000090700 | OBG//DRG2//BACTERIAL GTPASE |
| 9 | 0.0000090193 | TRIGGER FACTOR//NASCENT CHAIN//COTRANSLATIONAL FOLDING |
| 10 | 0.0000077715 | SUP35//TRANSLATION TERMINATION//URE3 |