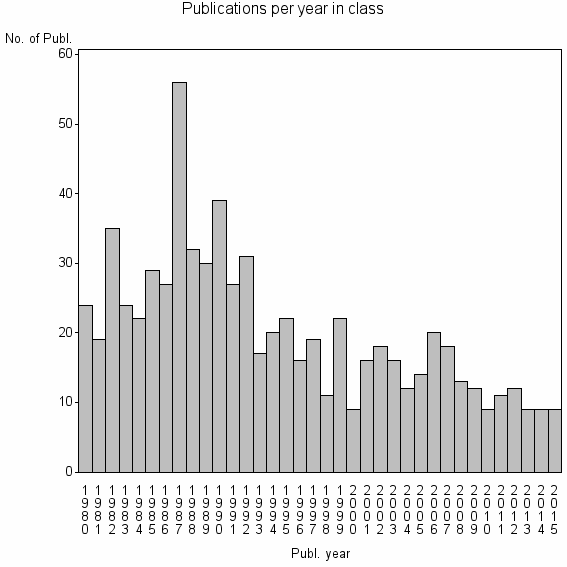
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 14068 | 729 | 34.2 | 65% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SD SEQUENCE | Author keyword | 13 | 80% | 1% | 8 |
| 2 | RIBOSOMAL PROTEIN S1 | Author keyword | 13 | 55% | 2% | 16 |
| 3 | SHINE DALGARNO SEQUENCE | Author keyword | 12 | 43% | 3% | 22 |
| 4 | LEADERLESS MRNA | Author keyword | 9 | 59% | 1% | 10 |
| 5 | GC RICH GENE | Author keyword | 4 | 75% | 0% | 3 |
| 6 | TRANSLATION INITIATION REGION | Author keyword | 4 | 41% | 1% | 7 |
| 7 | EPSILON SEQUENCE | Author keyword | 3 | 100% | 0% | 3 |
| 8 | FORMYLMETHIONYL TRANSFER RNA | Author keyword | 3 | 100% | 0% | 3 |
| 9 | LEADERLESS MRNAS | Author keyword | 3 | 100% | 0% | 3 |
| 10 | S1 RIBOSOMAL PROTEIN | Author keyword | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SD SEQUENCE | 13 | 80% | 1% | 8 | Search SD+SEQUENCE | Search SD+SEQUENCE |
| 2 | RIBOSOMAL PROTEIN S1 | 13 | 55% | 2% | 16 | Search RIBOSOMAL+PROTEIN+S1 | Search RIBOSOMAL+PROTEIN+S1 |
| 3 | SHINE DALGARNO SEQUENCE | 12 | 43% | 3% | 22 | Search SHINE+DALGARNO+SEQUENCE | Search SHINE+DALGARNO+SEQUENCE |
| 4 | LEADERLESS MRNA | 9 | 59% | 1% | 10 | Search LEADERLESS+MRNA | Search LEADERLESS+MRNA |
| 5 | GC RICH GENE | 4 | 75% | 0% | 3 | Search GC+RICH+GENE | Search GC+RICH+GENE |
| 6 | TRANSLATION INITIATION REGION | 4 | 41% | 1% | 7 | Search TRANSLATION+INITIATION+REGION | Search TRANSLATION+INITIATION+REGION |
| 7 | EPSILON SEQUENCE | 3 | 100% | 0% | 3 | Search EPSILON+SEQUENCE | Search EPSILON+SEQUENCE |
| 8 | FORMYLMETHIONYL TRANSFER RNA | 3 | 100% | 0% | 3 | Search FORMYLMETHIONYL+TRANSFER+RNA | Search FORMYLMETHIONYL+TRANSFER+RNA |
| 9 | LEADERLESS MRNAS | 3 | 100% | 0% | 3 | Search LEADERLESS+MRNAS | Search LEADERLESS+MRNAS |
| 10 | S1 RIBOSOMAL PROTEIN | 3 | 100% | 0% | 3 | Search S1+RIBOSOMAL+PROTEIN | Search S1+RIBOSOMAL+PROTEIN |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEIN S1 | 22 | 62% | 3% | 23 |
| 2 | DOWNSTREAM BOX | 15 | 45% | 4% | 26 |
| 3 | 5 TERMINAL START CODONS | 13 | 80% | 1% | 8 |
| 4 | RIBOSOME BINDING SITE | 11 | 33% | 4% | 27 |
| 5 | PENULTIMATE STEM | 9 | 83% | 1% | 5 |
| 6 | SHINE DALGARNO REGION | 9 | 55% | 2% | 11 |
| 7 | SHINE DALGARNO SEQUENCE | 8 | 33% | 3% | 19 |
| 8 | RIBOSOMAL PROTEIN S1 | 5 | 23% | 3% | 20 |
| 9 | LEADERLESS MESSENGER RNAS | 4 | 26% | 2% | 14 |
| 10 | RIBOSOME MESSENGER RECOGNITION | 4 | 75% | 0% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| A unified view of the initiation of protein synthesis | 2006 | 20 | 6 | 67% |
| Regulation of translation via mRNA structure in prokaryotes and eukaryotes | 2005 | 326 | 321 | 13% |
| Functional importance of RNA interactions in selection of translation initiation codons | 1997 | 51 | 53 | 34% |
| TRANSLATIONAL CONTROL OF PROKARYOTIC GENE-EXPRESSION | 1990 | 198 | 40 | 50% |
| Ribosome heterogeneity: another level of complexity in bacterial translation regulation | 2013 | 14 | 40 | 23% |
| PROKARYOTIC TRANSLATION - THE INTERACTIVE PATHWAY LEADING TO INITIATION | 1994 | 102 | 39 | 59% |
| Evolution and the universality of the mechanism of initiation of protein synthesis | 2009 | 19 | 18 | 33% |
| POSTTRANSCRIPTIONAL REGULATORY MECHANISMS IN ESCHERICHIA-COLI | 1988 | 547 | 83 | 23% |
| Initiation of eukaryotic and prokaryotic protein synthesis: A selective accessibility and multisubstrate enzyme reaction | 2007 | 3 | 12 | 58% |
| ENHANCED TRANSLATIONAL EFFICIENCY WITH 2-CISTRON EXPRESSION SYSTEM | 1990 | 49 | 16 | 81% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | G BONTCHEV | 1 | 50% | 0.1% | 1 |
| 2 | GENOPOLE PLATEAU PROTEOM | 1 | 50% | 0.1% | 1 |
| 3 | INTEGRATED BIOREFINERY | 1 | 50% | 0.1% | 1 |
| 4 | MODERN THEORET BIOL | 1 | 50% | 0.1% | 1 |
| 5 | SELECT IND MICROORGANISMS | 1 | 50% | 0.1% | 1 |
| 6 | CHEM CHEM ENZYMOL | 0 | 33% | 0.1% | 1 |
| 7 | DOMME | 0 | 33% | 0.1% | 1 |
| 8 | IND MICROBIOL BIOPROC | 0 | 33% | 0.1% | 1 |
| 9 | SORBONNE PARIS CITE BIOL PHYSICOCHIM | 0 | 33% | 0.1% | 1 |
| 10 | UPR 9073 CNRS | 0 | 33% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000173255 | Q BETA REPLICASE//POLONIES//RNA PHAGE |
| 2 | 0.0000166250 | EXPT BIOTECHNOL PLANT//HUMAN CONSENSUS INTERFERON ALPHA//A21 DESAMIDOINSULIN |
| 3 | 0.0000121819 | MRNA ANALOG//80S RIBOSOME//HUMAN RIBOSOME |
| 4 | 0.0000120897 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 5 | 0.0000113758 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 6 | 0.0000109218 | BACTERIOPHAGE T5//HOST PHAGE INTERACTION//LYTIC CONVERSION |
| 7 | 0.0000108336 | TRANS TRANSLATION//TMRNA//SMPB |
| 8 | 0.0000100529 | TRANSLATIONAL SELECTION//CODON USAGE//SYNONYMOUS CODON USAGE |
| 9 | 0.0000079485 | ERM METHYLTRANSFERASE//CMLA//MLS ANTIBIOTIC RESISTANCE |
| 10 | 0.0000074538 | HIGH CELL DENSITY CULTIVATION//RECOMBINANT ESCHERICHIA COLI//ACETATE EXCRETION |