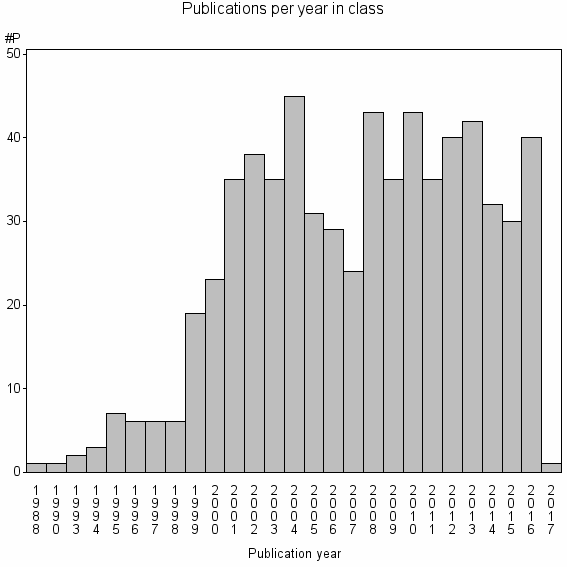
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 16463 | 652 | 53.5 | 94% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 16463 | 1 | MRNA EXPORT//NXF1//THO COMPLEX | 652 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MRNA EXPORT | authKW | 2061538 | 14% | 49% | 90 |
| 2 | NXF1 | authKW | 374649 | 2% | 67% | 12 |
| 3 | THO COMPLEX | authKW | 354161 | 2% | 69% | 11 |
| 4 | TREX | authKW | 337181 | 2% | 60% | 12 |
| 5 | MEX67P | authKW | 327825 | 1% | 100% | 7 |
| 6 | TREX COMPLEX | authKW | 299722 | 1% | 80% | 8 |
| 7 | MRNA NUCLEAR EXPORT | authKW | 292693 | 2% | 63% | 10 |
| 8 | TREX 2 | authKW | 272473 | 1% | 73% | 8 |
| 9 | SUB2 | authKW | 254972 | 1% | 78% | 7 |
| 10 | MEX67 | authKW | 240849 | 1% | 86% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Cell Biology | 7276 | 54% | 0% | 349 |
| 2 | Biochemistry & Molecular Biology | 3606 | 61% | 0% | 396 |
| 3 | Genetics & Heredity | 653 | 14% | 0% | 93 |
| 4 | Developmental Biology | 514 | 6% | 0% | 39 |
| 5 | Biophysics | 162 | 7% | 0% | 44 |
| 6 | Mycology | 15 | 1% | 0% | 5 |
| 7 | Virology | 8 | 1% | 0% | 9 |
| 8 | Biotechnology & Applied Microbiology | 5 | 3% | 0% | 18 |
| 9 | Biochemical Research Methods | 4 | 2% | 0% | 12 |
| 10 | Andrology | 2 | 0% | 0% | 1 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOMOL NETWORKS S | 176514 | 1% | 54% | 7 |
| 2 | GENE EXP S COUPLED RNA TRANSPORT | 140496 | 0% | 100% | 3 |
| 3 | CELL BIOL FRONTIERS GENET | 93664 | 0% | 100% | 2 |
| 4 | ANDALUZ BIOL MOL MED REGENERAT CABIMER | 51446 | 2% | 11% | 10 |
| 5 | TRANSCRIPT TORS | 49948 | 1% | 27% | 4 |
| 6 | HUMAN RETROVIRUS PATHOGENESIS SECT | 48617 | 1% | 12% | 9 |
| 7 | ABT ENTWICKLUNGSBIOCHEMIE | 46832 | 0% | 100% | 1 |
| 8 | BIOCHEM MOL HUMAN GENET | 46832 | 0% | 100% | 1 |
| 9 | CELL BIOL S7 214 | 46832 | 0% | 100% | 1 |
| 10 | CNRS UMR7592EQUIPE ELLISEE LIGUE CANC | 46832 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA-A PUBLICATION OF THE RNA SOCIETY | 14699 | 3% | 2% | 20 |
| 2 | NUCLEUS-AUSTIN | 10206 | 1% | 4% | 6 |
| 3 | RNA | 7362 | 3% | 1% | 20 |
| 4 | MOLECULAR CELL | 4916 | 4% | 0% | 24 |
| 5 | GENES & DEVELOPMENT | 4903 | 4% | 0% | 28 |
| 6 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 4453 | 2% | 1% | 10 |
| 7 | MOLECULAR AND CELLULAR BIOLOGY | 4033 | 7% | 0% | 44 |
| 8 | MOLECULAR BIOLOGY OF THE CELL | 2947 | 4% | 0% | 23 |
| 9 | EMBO JOURNAL | 2768 | 5% | 0% | 32 |
| 10 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 2034 | 1% | 1% | 4 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MRNA EXPORT | 2061538 | 14% | 49% | 90 | Search MRNA+EXPORT | Search MRNA+EXPORT |
| 2 | NXF1 | 374649 | 2% | 67% | 12 | Search NXF1 | Search NXF1 |
| 3 | THO COMPLEX | 354161 | 2% | 69% | 11 | Search THO+COMPLEX | Search THO+COMPLEX |
| 4 | TREX | 337181 | 2% | 60% | 12 | Search TREX | Search TREX |
| 5 | MEX67P | 327825 | 1% | 100% | 7 | Search MEX67P | Search MEX67P |
| 6 | TREX COMPLEX | 299722 | 1% | 80% | 8 | Search TREX+COMPLEX | Search TREX+COMPLEX |
| 7 | MRNA NUCLEAR EXPORT | 292693 | 2% | 63% | 10 | Search MRNA+NUCLEAR+EXPORT | Search MRNA+NUCLEAR+EXPORT |
| 8 | TREX 2 | 272473 | 1% | 73% | 8 | Search TREX+2 | Search TREX+2 |
| 9 | SUB2 | 254972 | 1% | 78% | 7 | Search SUB2 | Search SUB2 |
| 10 | MEX67 | 240849 | 1% | 86% | 6 | Search MEX67 | Search MEX67 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | VALKOV, E , DEAN, JC , JANI, D , KUHLMANN, SI , STEWART, M , (2012) STRUCTURAL BASIS FOR THE ASSEMBLY AND DISASSEMBLY OF MRNA NUCLEAR EXPORT COMPLEXES.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1819. ISSUE 6. P. 578-592 | 101 | 71% | 13 |
| 2 | KATAHIRA, J , (2015) NUCLEAR EXPORT OF MESSENGER RNA.GENES. VOL. 6. ISSUE 2. P. 163 -184 | 101 | 63% | 6 |
| 3 | HEATH, CG , VIPHAKONE, N , WILSON, SA , (2016) THE ROLE OF TREX IN GENE EXPRESSION AND DISEASE.BIOCHEMICAL JOURNAL. VOL. 473. ISSUE . P. 2911 -2935 | 112 | 57% | 0 |
| 4 | NINO, CA , HERISSANT, L , BABOUR, A , DARGEMONT, C , (2013) MRNA NUCLEAR EXPORT IN YEAST.CHEMICAL REVIEWS. VOL. 113. ISSUE 11. P. 8523 -8545 | 125 | 43% | 24 |
| 5 | WICKRAMASINGHE, VO , LASKEY, RA , (2015) CONTROL OF MAMMALIAN GENE EXPRESSION BY SELECTIVE MRNA EXPORT.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 16. ISSUE 7. P. 431 -442 | 80 | 53% | 17 |
| 6 | KATAHIRA, J , (2012) MRNA EXPORT AND THE TREX COMPLEX.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1819. ISSUE 6. P. 507-513 | 71 | 72% | 30 |
| 7 | ROUGEMAILLE, M , VILLA, T , GUDIPATI, RK , LIBRIL, D , (2008) MRNA JOURNEY TO THE CYTOPLASM: ATTIRE REQUIRED.BIOLOGY OF THE CELL. VOL. 100. ISSUE 6. P. 327-342 | 85 | 66% | 17 |
| 8 | KELLY, SM , CORBETT, AH , (2009) MESSENGER RNA EXPORT FROM THE NUCLEUS: A SERIES OF MOLECULAR WARDROBE CHANGES.TRAFFIC. VOL. 10. ISSUE 9. P. 1199 -1208 | 70 | 72% | 42 |
| 9 | ERKMANN, JA , KUTAY, U , (2004) NUCLEAR EXPORT OF MRNA: FROM THE SITE OF TRANSCRIPTION TO THE CYTOPLASM.EXPERIMENTAL CELL RESEARCH. VOL. 296. ISSUE 1. P. 12-20 | 67 | 78% | 87 |
| 10 | PALAZZO, AF , TRUONG, M , (2016) SINGLE PARTICLE IMAGING OF MRNAS CROSSING THE NUCLEAR PORE: SURFING ON THE EDGE.BIOESSAYS. VOL. 38. ISSUE 8. P. 744 -750 | 57 | 66% | 0 |
Classes with closest relation at Level 1 |