Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
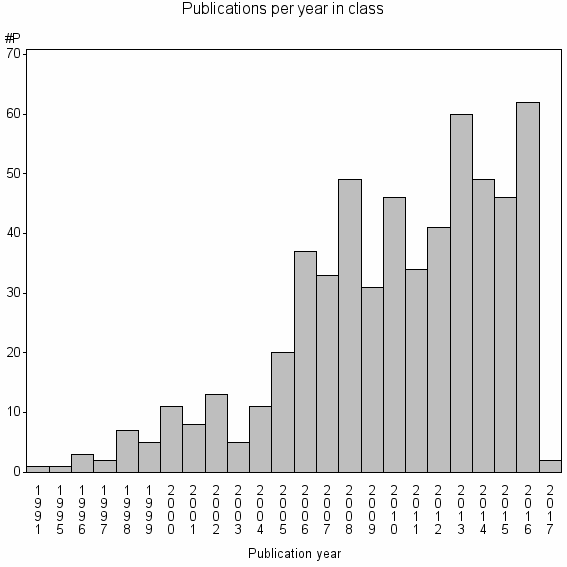
| 17891 | 577 | 51.5 | 95% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 17891 | 1 | RRP6//MRNP BIOGENESIS METAB//DIS3 | 577 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RRP6 | authKW | 804933 | 3% | 89% | 17 |
| 2 | MRNP BIOGENESIS METAB | address | 737010 | 6% | 41% | 34 |
| 3 | DIS3 | authKW | 695165 | 3% | 77% | 17 |
| 4 | NUCLEAR EXOSOME | authKW | 423358 | 1% | 100% | 8 |
| 5 | RNA EXOSOME | authKW | 423350 | 2% | 67% | 12 |
| 6 | RNA SURVEILLANCE | authKW | 255510 | 2% | 37% | 13 |
| 7 | RNA DEGRADATION | authKW | 233261 | 6% | 13% | 33 |
| 8 | DIS3L2 | authKW | 220497 | 1% | 83% | 5 |
| 9 | EXORIBONUCLEASE | authKW | 175916 | 3% | 21% | 16 |
| 10 | NAB3 | authKW | 169341 | 1% | 80% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 4688 | 72% | 0% | 418 |
| 2 | Cell Biology | 4128 | 43% | 0% | 250 |
| 3 | Genetics & Heredity | 589 | 14% | 0% | 83 |
| 4 | Biophysics | 172 | 7% | 0% | 42 |
| 5 | Developmental Biology | 122 | 3% | 0% | 19 |
| 6 | Biology | 48 | 3% | 0% | 19 |
| 7 | Biochemical Research Methods | 42 | 4% | 0% | 22 |
| 8 | Mycology | 39 | 1% | 0% | 7 |
| 9 | Biotechnology & Applied Microbiology | 36 | 5% | 0% | 28 |
| 10 | Medicine, Research & Experimental | 6 | 2% | 0% | 14 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MRNP BIOGENESIS METAB | 737010 | 6% | 41% | 34 |
| 2 | UNITE GENET INTERACT MACROMOL | 116777 | 1% | 28% | 8 |
| 3 | PEKING TSINGHUA BIOL SCI | 105839 | 0% | 100% | 2 |
| 4 | RNA BIOL GENOME MED | 70558 | 0% | 67% | 2 |
| 5 | STRUCT CELL BIOL | 67371 | 3% | 8% | 16 |
| 6 | WELLCOME TRUST CELL BIOL | 56067 | 5% | 4% | 29 |
| 7 | BEIJING ADV INNOVAT STRUCT BIOL LIFE SCI | 52920 | 0% | 100% | 1 |
| 8 | BIOINFORMAT COMPUTAT SYST BIOL SUMMER | 52920 | 0% | 100% | 1 |
| 9 | BIOMOL UTRECHT PHARMACEUT SCI | 52920 | 0% | 100% | 1 |
| 10 | C T PROGRAM GENE MECHSAKYO KU | 52920 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR CELL | 14732 | 7% | 1% | 39 |
| 2 | RNA | 14083 | 5% | 1% | 26 |
| 3 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 7052 | 1% | 2% | 7 |
| 4 | EMBO REPORTS | 4776 | 2% | 1% | 14 |
| 5 | RNA-A PUBLICATION OF THE RNA SOCIETY | 4144 | 2% | 1% | 10 |
| 6 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 4119 | 2% | 1% | 13 |
| 7 | RNA BIOLOGY | 2814 | 1% | 1% | 7 |
| 8 | NUCLEIC ACIDS RESEARCH | 2666 | 8% | 0% | 45 |
| 9 | MOLECULAR AND CELLULAR BIOLOGY | 2105 | 5% | 0% | 30 |
| 10 | PLOS GENETICS | 1955 | 3% | 0% | 15 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RRP6 | 804933 | 3% | 89% | 17 | Search RRP6 | Search RRP6 |
| 2 | DIS3 | 695165 | 3% | 77% | 17 | Search DIS3 | Search DIS3 |
| 3 | NUCLEAR EXOSOME | 423358 | 1% | 100% | 8 | Search NUCLEAR+EXOSOME | Search NUCLEAR+EXOSOME |
| 4 | RNA EXOSOME | 423350 | 2% | 67% | 12 | Search RNA+EXOSOME | Search RNA+EXOSOME |
| 5 | RNA SURVEILLANCE | 255510 | 2% | 37% | 13 | Search RNA+SURVEILLANCE | Search RNA+SURVEILLANCE |
| 6 | RNA DEGRADATION | 233261 | 6% | 13% | 33 | Search RNA+DEGRADATION | Search RNA+DEGRADATION |
| 7 | DIS3L2 | 220497 | 1% | 83% | 5 | Search DIS3L2 | Search DIS3L2 |
| 8 | EXORIBONUCLEASE | 175916 | 3% | 21% | 16 | Search EXORIBONUCLEASE | Search EXORIBONUCLEASE |
| 9 | NAB3 | 169341 | 1% | 80% | 4 | Search NAB3 | Search NAB3 |
| 10 | TRAMP COMPLEX | 169341 | 1% | 80% | 4 | Search TRAMP+COMPLEX | Search TRAMP+COMPLEX |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | KILCHERT, C , WITTMANN, S , VASILJEVA, L , (2016) THE REGULATION AND FUNCTIONS OF THE NUCLEAR RNA EXOSOME COMPLEX.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 17. ISSUE 4. P. 227 -239 | 87 | 54% | 11 |
| 2 | CHLEBOWSKI, A , LUBAS, M , JENSEN, TH , DZIEMBOWSKI, A , (2013) RNA DECAY MACHINES: THE EXOSOME.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1829. ISSUE 6-7. P. 552 -560 | 72 | 74% | 68 |
| 3 | SCHNEIDER, C , TOLLERVEY, D , (2013) THREADING THE BARREL OF THE RNA EXOSOME.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 38. ISSUE 10. P. 485-493 | 68 | 85% | 36 |
| 4 | HAN, J , VAN HOOF, A , (2016) THE RNA EXOSOME CHANNELING AND DIRECT ACCESS CONFORMATIONS HAVE DISTINCT IN VIVO FUNCTIONS.CELL REPORTS. VOL. 16. ISSUE 12. P. 3348 -3358 | 54 | 81% | 1 |
| 5 | FOX, MJ , MOSLEY, AL , (2016) RRP6: INTEGRATED ROLES IN NUCLEAR RNA METABOLISM AND TRANSCRIPTION TERMINATION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 7. ISSUE 1. P. 91 -104 | 57 | 76% | 1 |
| 6 | SCHMIDT, K , BUTLER, JS , (2013) NUCLEAR RNA SURVEILLANCE: ROLE OF TRAMP IN CONTROLLING EXOSOME SPECIFICITY.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 4. ISSUE 2. P. 217-231 | 69 | 61% | 28 |
| 7 | MITCHELL, P , (2014) EXOSOME SUBSTRATE TARGETING: THE LONG AND SHORT OF IT.BIOCHEMICAL SOCIETY TRANSACTIONS. VOL. 42. ISSUE . P. 1129 -1134 | 52 | 85% | 7 |
| 8 | JANUSZYK, K , LIMA, CD , (2014) THE EUKARYOTIC RNA EXOSOME.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 24. ISSUE . P. 132-140 | 45 | 80% | 29 |
| 9 | KISS, DL , ANDRULIS, ED , (2011) THE EXOZYME MODEL: A CONTINUUM OF FUNCTIONALLY DISTINCT COMPLEXES.RNA. VOL. 17. ISSUE 1. P. 1-13 | 71 | 64% | 16 |
| 10 | SCHUCH, B , FEIGENBUTZ, M , MAKINO, DL , FALK, S , BASQUIN, C , MITCHELL, P , CONTI, E , (2014) THE EXOSOME-BINDING FACTORS RRP6 AND RRP47 FORM A COMPOSITE SURFACE FOR RECRUITING THE MTR4 HELICASE.EMBO JOURNAL. VOL. 33. ISSUE 23. P. 2829 -2846 | 51 | 78% | 11 |
Classes with closest relation at Level 1 |