Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
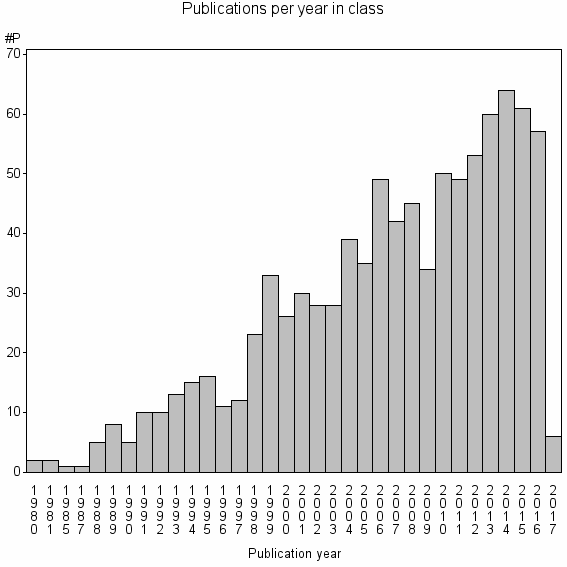
| 12218 | 923 | 48.5 | 91% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 12218 | 1 | RNA HELICASE//DEAD BOX PROTEIN//DEAD BOX | 923 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA HELICASE | authKW | 1437063 | 13% | 36% | 121 |
| 2 | DEAD BOX PROTEIN | authKW | 1037378 | 6% | 56% | 56 |
| 3 | DEAD BOX | authKW | 1001014 | 5% | 64% | 47 |
| 4 | DDX3 | authKW | 766480 | 4% | 70% | 33 |
| 5 | DEAD BOX RNA HELICASE | authKW | 604887 | 3% | 57% | 32 |
| 6 | RNA UNWINDING | authKW | 381402 | 2% | 82% | 14 |
| 7 | P68 RNA HELICASE | authKW | 340261 | 1% | 86% | 12 |
| 8 | UNWINDING ENZYME | authKW | 328860 | 1% | 76% | 13 |
| 9 | P68 | authKW | 315228 | 2% | 53% | 18 |
| 10 | DEAD BOX HELICASE | authKW | 199655 | 1% | 46% | 13 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 5775 | 64% | 0% | 593 |
| 2 | Cell Biology | 1816 | 24% | 0% | 220 |
| 3 | Biophysics | 465 | 9% | 0% | 84 |
| 4 | Genetics & Heredity | 353 | 9% | 0% | 87 |
| 5 | Oncology | 230 | 10% | 0% | 94 |
| 6 | Virology | 119 | 3% | 0% | 30 |
| 7 | Plant Sciences | 105 | 6% | 0% | 58 |
| 8 | Biochemical Research Methods | 61 | 4% | 0% | 34 |
| 9 | Developmental Biology | 56 | 2% | 0% | 18 |
| 10 | Microbiology | 36 | 4% | 0% | 38 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SECT MOL ENDOCRINOL | 153955 | 2% | 29% | 16 |
| 2 | RNA MOL BIOL | 66254 | 3% | 7% | 30 |
| 3 | HAEMATOPATHOL | 58847 | 1% | 16% | 11 |
| 4 | BIOCHEM CRC CELL TRANSFORMAT | 33081 | 0% | 100% | 1 |
| 5 | BIOSCI BIOECHNOL | 33081 | 0% | 100% | 1 |
| 6 | CHILD NEUROL PALLIAT CARE | 33081 | 0% | 100% | 1 |
| 7 | CNRS BIOL MODELLING CELLUMR 5239U1210 | 33081 | 0% | 100% | 1 |
| 8 | ECOLE DOCTORALE GGC 426 | 33081 | 0% | 100% | 1 |
| 9 | ELE ON MICROSCOPY MOL CYTOL | 33081 | 0% | 100% | 1 |
| 10 | ELECT MICROSCOPY MOL CYTOL | 33081 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA BIOLOGY | 5171 | 1% | 1% | 12 |
| 2 | NUCLEIC ACIDS RESEARCH | 3912 | 7% | 0% | 69 |
| 3 | RNA-A PUBLICATION OF THE RNA SOCIETY | 1650 | 1% | 1% | 8 |
| 4 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 1534 | 1% | 1% | 7 |
| 5 | JOURNAL OF MOLECULAR BIOLOGY | 1318 | 3% | 0% | 31 |
| 6 | RNA | 1287 | 1% | 0% | 10 |
| 7 | KOREAN JOURNAL OF BIOCHEMISTRY | 1065 | 0% | 3% | 1 |
| 8 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 805 | 0% | 1% | 3 |
| 9 | GENE | 792 | 2% | 0% | 22 |
| 10 | JOURNAL OF BIOLOGICAL CHEMISTRY | 728 | 7% | 0% | 61 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA HELICASE | 1437063 | 13% | 36% | 121 | Search RNA+HELICASE | Search RNA+HELICASE |
| 2 | DEAD BOX PROTEIN | 1037378 | 6% | 56% | 56 | Search DEAD+BOX+PROTEIN | Search DEAD+BOX+PROTEIN |
| 3 | DEAD BOX | 1001014 | 5% | 64% | 47 | Search DEAD+BOX | Search DEAD+BOX |
| 4 | DDX3 | 766480 | 4% | 70% | 33 | Search DDX3 | Search DDX3 |
| 5 | DEAD BOX RNA HELICASE | 604887 | 3% | 57% | 32 | Search DEAD+BOX+RNA+HELICASE | Search DEAD+BOX+RNA+HELICASE |
| 6 | RNA UNWINDING | 381402 | 2% | 82% | 14 | Search RNA+UNWINDING | Search RNA+UNWINDING |
| 7 | P68 RNA HELICASE | 340261 | 1% | 86% | 12 | Search P68+RNA+HELICASE | Search P68+RNA+HELICASE |
| 8 | UNWINDING ENZYME | 328860 | 1% | 76% | 13 | Search UNWINDING+ENZYME | Search UNWINDING+ENZYME |
| 9 | P68 | 315228 | 2% | 53% | 18 | Search P68 | Search P68 |
| 10 | DEAD BOX HELICASE | 199655 | 1% | 46% | 13 | Search DEAD+BOX+HELICASE | Search DEAD+BOX+HELICASE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PUTNAM, AA , JANKOWSKY, E , (2013) DEAD-BOX HELICASES AS INTEGRATORS OF RNA, NUCLEOTIDE AND PROTEIN BINDING.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1829. ISSUE 8. P. 884-893 | 77 | 71% | 32 |
| 2 | HILBERT, M , KAROW, AR , KLOSTERMEIER, D , (2009) THE MECHANISM OF ATP-DEPENDENT RNA UNWINDING BY DEAD BOX PROTEINS.BIOLOGICAL CHEMISTRY. VOL. 390. ISSUE 12. P. 1237 -1250 | 79 | 77% | 48 |
| 3 | LINDER, P , JANKOWSKY, E , (2011) FROM UNWINDING TO CLAMPING - THE DEAD BOX RNA HELICASE FAMILY.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 12. ISSUE 8. P. 505 -516 | 72 | 53% | 240 |
| 4 | CORDIN, O , BANROQUES, J , TANNER, NK , LINDER, P , (2006) THE DEAD-BOX PROTEIN FAMILY OF RNA HELICASES.GENE. VOL. 367. ISSUE . P. 17 -37 | 79 | 52% | 497 |
| 5 | RUDOLPH, MG , KLOSTERMEIER, D , (2015) WHEN CORE COMPETENCE IS NOT ENOUGH: FUNCTIONAL INTERPLAY OF THE DEAD-BOX HELICASE CORE WITH ANCILLARY DOMAINS AND AUXILIARY FACTORS IN RNA BINDING AND UNWINDING.BIOLOGICAL CHEMISTRY. VOL. 396. ISSUE 8. P. 849 -865 | 61 | 88% | 2 |
| 6 | JARMOSKAITE, I , RUSSELL, R , (2011) DEAD-BOX PROTEINS AS RNA HELICASES AND CHAPERONES.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 2. ISSUE 1. P. 135-152 | 75 | 69% | 48 |
| 7 | JARMOSKAITE, I , RUSSELL, R , (2014) RNA HELICASE PROTEINS AS CHAPERONES AND REMODELERS.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 83. VOL. 83. ISSUE . P. 697 -725 | 97 | 42% | 36 |
| 8 | SARKAR, M , GHOSH, MK , (2016) DEAD BOX RNA HELICASES: CRUCIAL REGULATORS OF GENE EXPRESSION AND ONCOGENESIS.FRONTIERS IN BIOSCIENCE-LANDMARK. VOL. 21. ISSUE . P. 225 -250 | 93 | 53% | 0 |
| 9 | FULLER-PACE, FV , (2013) THE DEAD BOX PROTEINS DDX5 (P68) AND DDX17 (P72): MULTI-TASKING TRANSCRIPTIONAL REGULATORS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1829. ISSUE 8. P. 756-763 | 53 | 84% | 26 |
| 10 | SHARMA, D , JANKOWSKY, E , (2014) THE DED1/DDX3 SUBFAMILY OF DEAD-BOX RNA HELICASES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 49. ISSUE 4. P. 343 -360 | 84 | 51% | 18 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 13276 | UVRB//UVRD//UVRABC |
| 2 | 16463 | MRNA EXPORT//NXF1//THO COMPLEX |
| 3 | 3885 | EIF4E//EIF4F//CAP ANALOG |
| 4 | 3668 | SPLICEOSOME//PRE MRNA SPLICING//SNRNP |
| 5 | 19925 | VASA//NANAE F H WATER//PRIMORDIAL GERM CELLS |
| 6 | 6198 | P BODIES//STRESS GRANULES//DECAPPING |
| 7 | 15616 | COLD SHOCK PROTEIN//GLYCINE RICH RNA BINDING PROTEIN//RNA CHAPERONE |
| 8 | 10335 | NMD//UPF1//NONSENSE MEDIATED MRNA DECAY |
| 9 | 4461 | SNORNA//SMALL NUCLEOLAR RNA//PRE RRNA PROCESSING |
| 10 | 12194 | REV//RRE//REV PROTEIN |