Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
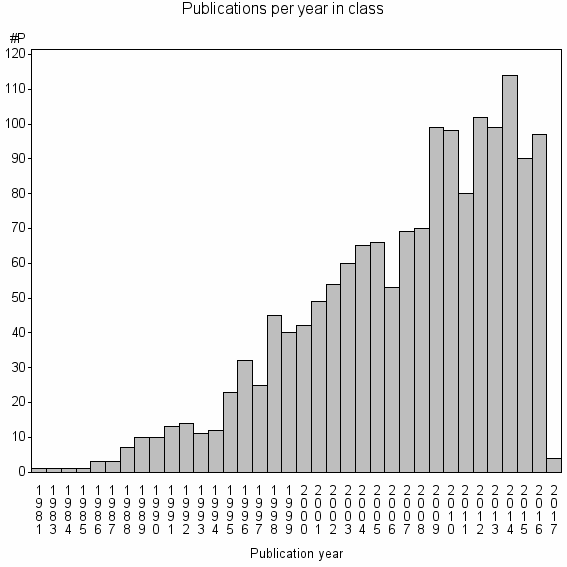
| 5889 | 1563 | 54.8 | 94% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 5889 | 1 | P TEFB//CDK9//RNA POLYMERASE II | 1563 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | P TEFB | authKW | 1315620 | 6% | 71% | 95 |
| 2 | CDK9 | authKW | 666619 | 4% | 58% | 59 |
| 3 | RNA POLYMERASE II | authKW | 578229 | 10% | 19% | 158 |
| 4 | HEXIM1 | authKW | 440174 | 2% | 87% | 26 |
| 5 | TRANSCRIPTION ELONGATION | authKW | 431857 | 4% | 34% | 66 |
| 6 | DSIF | authKW | 253159 | 1% | 72% | 18 |
| 7 | CTD | authKW | 218200 | 2% | 31% | 36 |
| 8 | CTD PHOSPHATASE | authKW | 206330 | 1% | 81% | 13 |
| 9 | CYCLIN T1 | authKW | 205093 | 1% | 50% | 21 |
| 10 | CYCLIN T | authKW | 157574 | 1% | 73% | 11 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Cell Biology | 13713 | 48% | 0% | 746 |
| 2 | Biochemistry & Molecular Biology | 10424 | 66% | 0% | 1034 |
| 3 | Genetics & Heredity | 1863 | 15% | 0% | 241 |
| 4 | Developmental Biology | 1276 | 6% | 0% | 95 |
| 5 | Biophysics | 374 | 7% | 0% | 104 |
| 6 | Virology | 194 | 3% | 0% | 50 |
| 7 | Biology | 44 | 2% | 0% | 35 |
| 8 | Biochemical Research Methods | 40 | 3% | 0% | 42 |
| 9 | Biotechnology & Applied Microbiology | 28 | 3% | 0% | 53 |
| 10 | Mycology | 22 | 1% | 0% | 10 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ROSALIND RUSSELL MED | 139508 | 1% | 36% | 20 |
| 2 | REGULAT EXP S GENET | 79761 | 0% | 58% | 7 |
| 3 | UMR 8541 | 69523 | 1% | 20% | 18 |
| 4 | EXP S ENGN GRP | 62501 | 1% | 40% | 8 |
| 5 | EDINBURGH SYST BIOL | 58604 | 0% | 100% | 3 |
| 6 | GRP PHYS BIOCHEM | 48832 | 0% | 50% | 5 |
| 7 | GENET MOL GEN BIOL | 44392 | 0% | 45% | 5 |
| 8 | IFIBYNE UBA CONICET | 43952 | 0% | 75% | 3 |
| 9 | ISRAEL PERSONALIZED MED INCPM | 39069 | 0% | 100% | 2 |
| 10 | MOL GENET BLOOD DEV | 39069 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR CELL | 22846 | 5% | 1% | 80 |
| 2 | GENES & DEVELOPMENT | 13535 | 5% | 1% | 72 |
| 3 | MOLECULAR AND CELLULAR BIOLOGY | 13404 | 8% | 1% | 124 |
| 4 | NATURE STRUCTURAL & MOLECULAR BIOLOGY | 8088 | 2% | 1% | 30 |
| 5 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 5303 | 1% | 3% | 10 |
| 6 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 3629 | 1% | 1% | 14 |
| 7 | RNA | 3362 | 1% | 1% | 21 |
| 8 | JOURNAL OF BIOLOGICAL CHEMISTRY | 2920 | 10% | 0% | 155 |
| 9 | NUCLEIC ACIDS RESEARCH | 2604 | 5% | 0% | 74 |
| 10 | RNA BIOLOGY | 2111 | 1% | 1% | 10 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | P TEFB | 1315620 | 6% | 71% | 95 | Search P+TEFB | Search P+TEFB |
| 2 | CDK9 | 666619 | 4% | 58% | 59 | Search CDK9 | Search CDK9 |
| 3 | RNA POLYMERASE II | 578229 | 10% | 19% | 158 | Search RNA+POLYMERASE+II | Search RNA+POLYMERASE+II |
| 4 | HEXIM1 | 440174 | 2% | 87% | 26 | Search HEXIM1 | Search HEXIM1 |
| 5 | TRANSCRIPTION ELONGATION | 431857 | 4% | 34% | 66 | Search TRANSCRIPTION+ELONGATION | Search TRANSCRIPTION+ELONGATION |
| 6 | DSIF | 253159 | 1% | 72% | 18 | Search DSIF | Search DSIF |
| 7 | CTD | 218200 | 2% | 31% | 36 | Search CTD | Search CTD |
| 8 | CTD PHOSPHATASE | 206330 | 1% | 81% | 13 | Search CTD+PHOSPHATASE | Search CTD+PHOSPHATASE |
| 9 | CYCLIN T1 | 205093 | 1% | 50% | 21 | Search CYCLIN+T1 | Search CYCLIN+T1 |
| 10 | CYCLIN T | 157574 | 1% | 73% | 11 | Search CYCLIN+T | Search CYCLIN+T |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | EICK, D , GEYER, M , (2013) THE RNA POLYMERASE II CARBOXY-TERMINAL DOMAIN (CTD) CODE.CHEMICAL REVIEWS. VOL. 113. ISSUE 11. P. 8456-8490 | 197 | 54% | 76 |
| 2 | HSIN, JP , MANLEY, JL , (2012) THE RNA POLYMERASE II CTD COORDINATES TRANSCRIPTION AND RNA PROCESSING.GENES & DEVELOPMENT. VOL. 26. ISSUE 19. P. 2119-2137 | 145 | 57% | 153 |
| 3 | JERONIMO, C , BATAILLE, AR , ROBERT, F , (2013) THE WRITERS, READERS, AND FUNCTIONS OF THE RNA POLYMERASE II C-TERMINAL DOMAIN CODE.CHEMICAL REVIEWS. VOL. 113. ISSUE 11. P. 8491-8522 | 210 | 48% | 22 |
| 4 | ZHOU, Q , LI, TD , PRICE, DH , (2012) RNA POLYMERASE II ELONGATION CONTROL.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 81. VOL. 81. ISSUE . P. 119-143 | 109 | 66% | 155 |
| 5 | CORDEN, JL , (2013) RNA POLYMERASE II C-TERMINAL DOMAIN: TETHERING TRANSCRIPTION TO TRANSCRIPT AND TEMPLATE.CHEMICAL REVIEWS. VOL. 113. ISSUE 11. P. 8423 -8455 | 221 | 41% | 34 |
| 6 | BENTLEY, DL , (2014) COUPLING MRNA PROCESSING WITH TRANSCRIPTION IN TIME AND SPACE.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 3. P. 163-175 | 104 | 57% | 117 |
| 7 | GUO, JN , PRICE, DH , (2013) RNA POLYMERASE II TRANSCRIPTION ELONGATION CONTROL.CHEMICAL REVIEWS. VOL. 113. ISSUE 11. P. 8583 -8603 | 188 | 49% | 28 |
| 8 | JERONIMO, C , COLLIN, P , ROBERT, F , (2016) THE RNA POLYMERASE II CTD: THE INCREASING COMPLEXITY OF A LOW-COMPLEXITY PROTEIN DOMAIN.JOURNAL OF MOLECULAR BIOLOGY. VOL. 428. ISSUE 12. P. 2607 -2622 | 96 | 69% | 7 |
| 9 | MCNAMARA, RP , BACON, CW , D'ORSO, I , (2016) TRANSCRIPTION ELONGATION CONTROL BY THE 7SK SNRNP COMPLEX: RELEASING THE PAUSE.CELL CYCLE. VOL. 15. ISSUE 16. P. 2115 -2123 | 88 | 87% | 1 |
| 10 | QUARESMA, AJC , BUGAI, A , BARBORIC, M , (2016) CRACKING THE CONTROL OF RNA POLYMERASE II ELONGATION BY 7SK SNRNP AND P-TEFB.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 16. P. 7527 -7539 | 94 | 78% | 1 |
Classes with closest relation at Level 1 |
| Rank | Class id | link |
|---|---|---|
| 1 | 23606 | CAK//SPY1//CDK ACTIVATING KINASE |
| 2 | 36779 | CDK11P58//PITSLRE//CDK11 |
| 3 | 14125 | USP22//SET1//UBIQUITIN SPECIFIC PROTEASE 22 |
| 4 | 8466 | POLYADENYLATION//ALTERNATIVE POLYADENYLATION//POLYA POLYMERASE |
| 5 | 4242 | TAT//TAT PROTEIN//HIV 1 TAT |
| 6 | 13537 | MEDIATOR COMPLEX//CDK8//MED12 |
| 7 | 17891 | RRP6//MRNP BIOGENESIS METAB//DIS3 |
| 8 | 17211 | RPB4//RPB7//INTERSPECIFIC COMPLEMENTATION |
| 9 | 16463 | MRNA EXPORT//NXF1//THO COMPLEX |
| 10 | 32852 | POLYPYRIMIDINE TRACT BINDING PROTEIN ASSOCIATED SPLICING FACTOR//P54NRB//NONO |