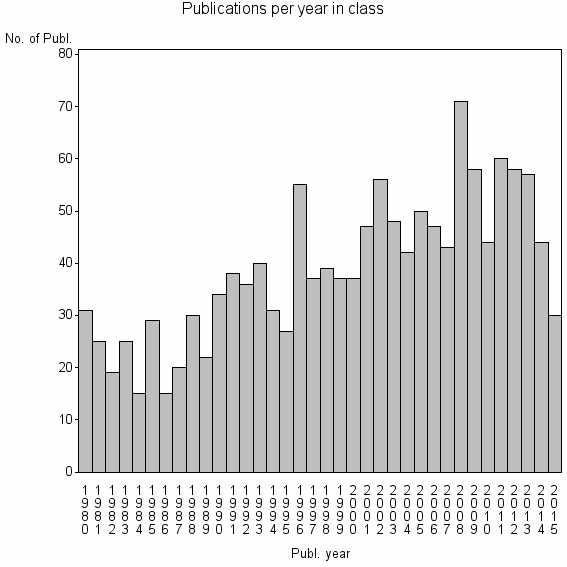
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 6587 | 1397 | 44.9 | 76% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RNASE E | Author keyword | 86 | 68% | 5% | 76 |
| 2 | RNASE G | Author keyword | 48 | 100% | 1% | 17 |
| 3 | POLYNUCLEOTIDE PHOSPHORYLASE | Author keyword | 40 | 66% | 3% | 37 |
| 4 | RNA DEGRADOSOME | Author keyword | 36 | 79% | 2% | 23 |
| 5 | DEGRADOSOME | Author keyword | 32 | 70% | 2% | 26 |
| 6 | PNPASE | Author keyword | 28 | 62% | 2% | 29 |
| 7 | RNASE II | Author keyword | 25 | 73% | 1% | 19 |
| 8 | RNA DEGRADATION | Author keyword | 16 | 25% | 4% | 57 |
| 9 | RNASE III | Author keyword | 14 | 37% | 2% | 30 |
| 10 | RIBONUCLEASE E | Author keyword | 11 | 100% | 0% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNASE E | 86 | 68% | 5% | 76 | Search RNASE+E | Search RNASE+E |
| 2 | RNASE G | 48 | 100% | 1% | 17 | Search RNASE+G | Search RNASE+G |
| 3 | POLYNUCLEOTIDE PHOSPHORYLASE | 40 | 66% | 3% | 37 | Search POLYNUCLEOTIDE+PHOSPHORYLASE | Search POLYNUCLEOTIDE+PHOSPHORYLASE |
| 4 | RNA DEGRADOSOME | 36 | 79% | 2% | 23 | Search RNA+DEGRADOSOME | Search RNA+DEGRADOSOME |
| 5 | DEGRADOSOME | 32 | 70% | 2% | 26 | Search DEGRADOSOME | Search DEGRADOSOME |
| 6 | PNPASE | 28 | 62% | 2% | 29 | Search PNPASE | Search PNPASE |
| 7 | RNASE II | 25 | 73% | 1% | 19 | Search RNASE+II | Search RNASE+II |
| 8 | RNA DEGRADATION | 16 | 25% | 4% | 57 | Search RNA+DEGRADATION | Search RNA+DEGRADATION |
| 9 | RNASE III | 14 | 37% | 2% | 30 | Search RNASE+III | Search RNASE+III |
| 10 | RIBONUCLEASE E | 11 | 100% | 0% | 6 | Search RIBONUCLEASE+E | Search RIBONUCLEASE+E |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RIBONUCLEASE E | 349 | 87% | 12% | 172 |
| 2 | POLYNUCLEOTIDE PHOSPHORYLASE | 189 | 56% | 16% | 228 |
| 3 | CAFA PROTEIN | 189 | 97% | 4% | 56 |
| 4 | RIBOSOMAL PROTEIN S20 | 118 | 95% | 3% | 39 |
| 5 | AMS GENE | 111 | 100% | 2% | 32 |
| 6 | DEGRADOSOME | 103 | 68% | 7% | 91 |
| 7 | RIBONUCLEASE II | 71 | 72% | 4% | 56 |
| 8 | RPSO MESSENGER RNA | 54 | 83% | 2% | 30 |
| 9 | MULTIPLE MUTANTS | 53 | 89% | 2% | 24 |
| 10 | ANTISENSE RNAI | 51 | 91% | 2% | 21 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| RNase E: at the interface of bacterial RNA processing and decay | 2013 | 42 | 117 | 82% |
| The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E | 2007 | 231 | 79 | 80% |
| The critical role of RNA processing and degradation in the control of gene expression | 2010 | 99 | 442 | 64% |
| RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases | 2012 | 33 | 78 | 77% |
| Endonucleolytic Initiation of mRNA Decay in Escherichia coli | 2009 | 64 | 200 | 77% |
| Maturation and Degradation of Ribosomal RNA in Bacteria | 2009 | 63 | 40 | 78% |
| The social fabric of the RNA degradosome | 2013 | 15 | 83 | 76% |
| Initiation of mRNA decay in bacteria | 2014 | 7 | 290 | 63% |
| Composition and conservation of the mRNA-degrading machinery in bacteria | 2011 | 18 | 72 | 96% |
| Degradation of mRNA in Escherichia coli: An old problem with some new twists | 1999 | 223 | 163 | 79% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | UPR 9073 | 8 | 23% | 2.2% | 31 |
| 2 | CNRS UPR 9073 | 6 | 53% | 0.6% | 8 |
| 3 | ICSN RMN | 3 | 57% | 0.3% | 4 |
| 4 | UMR 5100 | 3 | 16% | 1.1% | 16 |
| 5 | MIKRO MOL BIOL | 2 | 33% | 0.4% | 6 |
| 6 | GRP ARN | 2 | 27% | 0.4% | 6 |
| 7 | FRC550 | 2 | 43% | 0.2% | 3 |
| 8 | CNRS UPR9073 | 1 | 38% | 0.2% | 3 |
| 9 | SECURITE ALIMENTS MICROBIOL UMR1014 | 1 | 50% | 0.1% | 2 |
| 10 | UPR9073 | 1 | 13% | 0.6% | 9 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000203974 | TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME//TRNAHIS GUANYLYLTRANSFERASE |
| 2 | 0.0000173038 | HFQ//S RNA//RYHB |
| 3 | 0.0000144409 | EPIDEMIOLOGICAL SIGNIFICANCE//SIMILARITY IDENTIFICATION//AIRBORNE ESCHERICHIA COLI |
| 4 | 0.0000120897 | SD SEQUENCE//RIBOSOMAL PROTEIN S1//SHINE DALGARNO SEQUENCE |
| 5 | 0.0000119420 | MRNP BIOGENESIS METAB//DIS3//RRP6 |
| 6 | 0.0000116780 | TRANS TRANSLATION//TMRNA//SMPB |
| 7 | 0.0000107140 | RNASE P//RIBONUCLEASE P//CARTILAGE HAIR HYPOPLASIA |
| 8 | 0.0000067982 | COLD SHOCK PROTEIN//GLYCINE RICH RNA BINDING PROTEIN//CIRP |
| 9 | 0.0000065984 | OBG//DRG2//BACTERIAL GTPASE |
| 10 | 0.0000065431 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |