Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
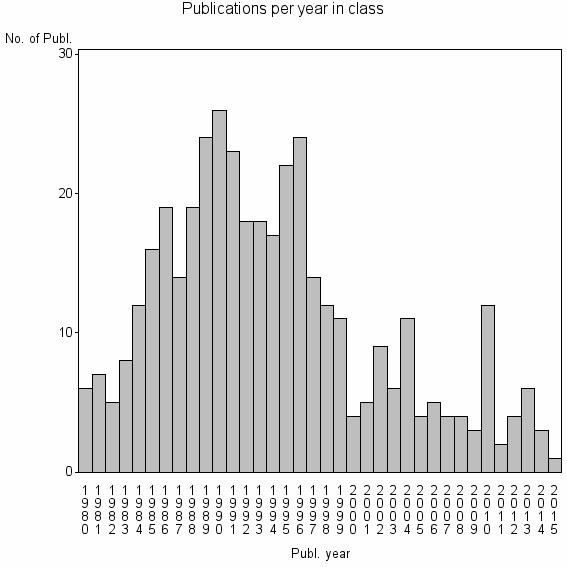
| 20449 | 398 | 28.9 | 57% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | Q BETA REPLICASE | Author keyword | 31 | 73% | 6% | 24 |
| 2 | POLONIES | Author keyword | 12 | 86% | 2% | 6 |
| 3 | RNA PHAGE | Author keyword | 9 | 67% | 2% | 8 |
| 4 | MOLECULAR COLONIES | Author keyword | 8 | 100% | 1% | 5 |
| 5 | SPONTANEOUS RNA SYNTHESIS | Author keyword | 8 | 100% | 1% | 5 |
| 6 | Q BETA PHAGE | Author keyword | 6 | 71% | 1% | 5 |
| 7 | BACTERIOPHAGE Q BETA | Author keyword | 4 | 42% | 2% | 8 |
| 8 | TEMPLATE RECOGNITION | Author keyword | 4 | 56% | 1% | 5 |
| 9 | MOLECULAR COLONY TECHNIQUE | Author keyword | 3 | 100% | 1% | 3 |
| 10 | RECOMBINANT MESSENGER RNA | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | Q BETA REPLICASE | 31 | 73% | 6% | 24 | Search Q+BETA+REPLICASE | Search Q+BETA+REPLICASE |
| 2 | POLONIES | 12 | 86% | 2% | 6 | Search POLONIES | Search POLONIES |
| 3 | RNA PHAGE | 9 | 67% | 2% | 8 | Search RNA+PHAGE | Search RNA+PHAGE |
| 4 | MOLECULAR COLONIES | 8 | 100% | 1% | 5 | Search MOLECULAR+COLONIES | Search MOLECULAR+COLONIES |
| 5 | SPONTANEOUS RNA SYNTHESIS | 8 | 100% | 1% | 5 | Search SPONTANEOUS+RNA+SYNTHESIS | Search SPONTANEOUS+RNA+SYNTHESIS |
| 6 | Q BETA PHAGE | 6 | 71% | 1% | 5 | Search Q+BETA+PHAGE | Search Q+BETA+PHAGE |
| 7 | BACTERIOPHAGE Q BETA | 4 | 42% | 2% | 8 | Search BACTERIOPHAGE+Q+BETA | Search BACTERIOPHAGE+Q+BETA |
| 8 | TEMPLATE RECOGNITION | 4 | 56% | 1% | 5 | Search TEMPLATE+RECOGNITION | Search TEMPLATE+RECOGNITION |
| 9 | MOLECULAR COLONY TECHNIQUE | 3 | 100% | 1% | 3 | Search MOLECULAR+COLONY+TECHNIQUE | Search MOLECULAR+COLONY+TECHNIQUE |
| 10 | RECOMBINANT MESSENGER RNA | 2 | 67% | 1% | 2 | Search RECOMBINANT+MESSENGER+RNA | Search RECOMBINANT+MESSENGER+RNA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BACTERIOPHAGE QBETA REPLICASE | 48 | 100% | 4% | 17 |
| 2 | QBETA REPLICASE | 15 | 45% | 7% | 26 |
| 3 | Q BETA REPLICASE | 14 | 30% | 10% | 40 |
| 4 | AUTOCATALYTIC SYNTHESIS | 11 | 100% | 2% | 6 |
| 5 | FREE RNA SYNTHESIS | 11 | 100% | 2% | 6 |
| 6 | RECOMBINANT RNA | 8 | 42% | 4% | 14 |
| 7 | PHAGE RNA | 6 | 80% | 1% | 4 |
| 8 | COLIPHAGE Q BETA | 5 | 55% | 2% | 6 |
| 9 | SELF ENCODED REPLICASE | 3 | 57% | 1% | 4 |
| 10 | RNA CHAIN | 3 | 100% | 1% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Molecular Colony Technique: A New Tool for Biomedical Research and Clinical Practice | 2008 | 3 | 96 | 32% |
| REPLICABLE RNA VECTORS - PROSPECTS FOR CELL-FREE GENE AMPLIFICATION, EXPRESSION, AND CLONING | 1995 | 13 | 60 | 60% |
| Nanocolonies: Detection, Cloning, and Analysis of Individual Molecules | 2008 | 2 | 110 | 31% |
| The puzzle of RNA recombination | 1999 | 31 | 33 | 27% |
| Omnipotent RNA | 2002 | 27 | 22 | 14% |
| Molecular evolution of RNA in vitro | 1997 | 18 | 71 | 34% |
| EXPONENTIAL AMPLIFICATION OF NUCLEIC-ACIDS - NEW DIAGNOSTICS USING DNA-POLYMERASES AND RNA REPLICASES | 1991 | 14 | 23 | 35% |
| Structures and Functions of Q beta Replicase: Translation Factors beyond Protein Synthesis | 2014 | 0 | 56 | 30% |
| The forces driving molecular evolution | 1998 | 4 | 91 | 26% |
| DARWINIAN SELECTION OF SELF-REPLICATING RNA MOLECULES | 1983 | 46 | 16 | 75% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL GENET MICROBIOL CANC TREATMENT | 1 | 50% | 0.3% | 1 |
| 2 | BIOCHEM KINET | 1 | 22% | 0.5% | 2 |
| 3 | INMABB | 0 | 13% | 0.5% | 2 |
| 4 | CALICIVIRUS | 0 | 17% | 0.3% | 1 |
| 5 | FORM FUNCT PROJECT | 0 | 17% | 0.3% | 1 |
| 6 | MATEMAT INMABB | 0 | 17% | 0.3% | 1 |
| 7 | QUIM ING QCA | 0 | 17% | 0.3% | 1 |
| 8 | YOMO DYNAM MICROSCALE REACT ENVIRONM PROJECT | 0 | 14% | 0.3% | 1 |
| 9 | CATEDRA GASTROENTEROL | 0 | 13% | 0.3% | 1 |
| 10 | MATEMAT BAHIA BLANCA | 0 | 10% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000173255 | SD SEQUENCE//RIBOSOMAL PROTEIN S1//SHINE DALGARNO SEQUENCE |
| 2 | 0.0000141529 | ERROR THRESHOLD//ERROR CATASTROPHE//LETHAL MUTAGENESIS |
| 3 | 0.0000106866 | RNA SECONDARY STRUCTURE//RNA SECONDARY STRUCTURE PREDICTION//RNA STRUCTURE PREDICTION |
| 4 | 0.0000091289 | TSITSIN MAIN BOT GARDENS//EARTHS PRIMARY AQUEOUS SHELL//REGULARITIES OF FUNCTIONING |
| 5 | 0.0000090802 | RNA WORLD//ORIGIN OF LIFE//ORIGINS OF LIFE AND EVOLUTION OF BIOSPHERES |
| 6 | 0.0000087365 | TOMBUSVIRIDAE//TOMBUSVIRUS//TYMV |
| 7 | 0.0000074423 | BIOINSPIRED NANOMAT//COWPEA CHLOROTIC MOTTLE VIRUS//INTEGRAT MOL BIOSCI |
| 8 | 0.0000072993 | GROUP I INTRON//SELF SPLICING//RIBOZYME |
| 9 | 0.0000065311 | PCR-METHODS AND APPLICATIONS//PLASMA VIREMIA//DIGOXIGENIN 11 DUTP |
| 10 | 0.0000052220 | EEUFMG//FUNEC//MULTI LAYER PERCEPTON |