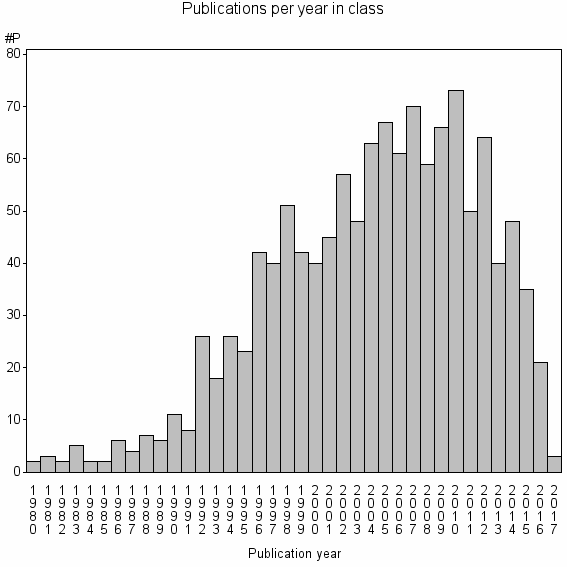
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8613 | 1236 | 31.4 | 71% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 79 | 3 | AGRONOMY//CROP SCIENCE//THEORETICAL AND APPLIED GENETICS | 80349 |
| 1130 | 2 | GENOMIC SELECTION//MAYDICA//GENOMIC PREDICTION | 9640 |
| 8613 | 1 | INTERVAL MAPPING//QTL MAPPING//GENETICS RESEARCH | 1236 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | INTERVAL MAPPING | authKW | 501894 | 3% | 47% | 43 |
| 2 | QTL MAPPING | authKW | 202811 | 6% | 10% | 80 |
| 3 | GENETICS RESEARCH | journal | 170668 | 6% | 9% | 74 |
| 4 | QUANTITATIVE TRAIT LOCI | authKW | 169105 | 9% | 6% | 113 |
| 5 | QTL DETECTION | authKW | 124132 | 1% | 36% | 14 |
| 6 | SELECTIVE GENOTYPING | authKW | 114960 | 1% | 29% | 16 |
| 7 | COURSE ENVIRONM MANAGEMENT SCI | address | 111162 | 0% | 75% | 6 |
| 8 | MULTIPLE QTL | authKW | 111162 | 0% | 75% | 6 |
| 9 | QTL | authKW | 106146 | 8% | 4% | 105 |
| 10 | ENDOSPERM TRAIT | authKW | 98813 | 0% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 32187 | 66% | 0% | 818 |
| 2 | Horticulture | 9026 | 14% | 0% | 175 |
| 3 | Agronomy | 4327 | 16% | 0% | 199 |
| 4 | Agriculture, Dairy & Animal Science | 4144 | 14% | 0% | 170 |
| 5 | Mathematical & Computational Biology | 3950 | 11% | 0% | 131 |
| 6 | Statistics & Probability | 1886 | 10% | 0% | 123 |
| 7 | Evolutionary Biology | 1598 | 8% | 0% | 97 |
| 8 | Plant Sciences | 1425 | 16% | 0% | 199 |
| 9 | Ecology | 352 | 7% | 0% | 87 |
| 10 | Biology | 270 | 5% | 0% | 60 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | COURSE ENVIRONM MANAGEMENT SCI | 111162 | 0% | 75% | 6 |
| 2 | SECT STAT GENOM | 86455 | 1% | 50% | 7 |
| 3 | STAT GENET | 71875 | 5% | 5% | 57 |
| 4 | BOT PLANT SCI | 66521 | 6% | 4% | 70 |
| 5 | FISHERIES OURCE ENVIRONM | 56461 | 0% | 57% | 4 |
| 6 | KEY FO T GENET GENE ENGN | 55581 | 0% | 75% | 3 |
| 7 | CIMMYT CHINA | 52305 | 0% | 35% | 6 |
| 8 | ORNAMENTAL GENET | 49407 | 0% | 100% | 2 |
| 9 | SECT STAT GENOM SOYBEAN IMPROVEMENT | 49407 | 0% | 100% | 2 |
| 10 | NUTZPFLANZENKUNDE | 44107 | 0% | 36% | 5 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENETICS RESEARCH | 170668 | 6% | 9% | 74 |
| 2 | GENETICS | 95588 | 18% | 2% | 222 |
| 3 | GENETICS SELECTION EVOLUTION | 81425 | 6% | 5% | 73 |
| 4 | THEORETICAL AND APPLIED GENETICS | 42451 | 11% | 1% | 131 |
| 5 | HEREDITY | 34938 | 6% | 2% | 80 |
| 6 | JOURNAL OF ANIMAL BREEDING AND GENETICS | 12412 | 2% | 2% | 25 |
| 7 | BMC GENETICS | 9937 | 2% | 2% | 26 |
| 8 | STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY | 9561 | 1% | 3% | 14 |
| 9 | BRIEFINGS IN BIOINFORMATICS | 6939 | 1% | 2% | 14 |
| 10 | GENETICAL RESEARCH | 5669 | 1% | 2% | 14 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | INTERVAL MAPPING | 501894 | 3% | 47% | 43 | Search INTERVAL+MAPPING | Search INTERVAL+MAPPING |
| 2 | QTL MAPPING | 202811 | 6% | 10% | 80 | Search QTL+MAPPING | Search QTL+MAPPING |
| 3 | QUANTITATIVE TRAIT LOCI | 169105 | 9% | 6% | 113 | Search QUANTITATIVE+TRAIT+LOCI | Search QUANTITATIVE+TRAIT+LOCI |
| 4 | QTL DETECTION | 124132 | 1% | 36% | 14 | Search QTL+DETECTION | Search QTL+DETECTION |
| 5 | SELECTIVE GENOTYPING | 114960 | 1% | 29% | 16 | Search SELECTIVE+GENOTYPING | Search SELECTIVE+GENOTYPING |
| 6 | MULTIPLE QTL | 111162 | 0% | 75% | 6 | Search MULTIPLE+QTL | Search MULTIPLE+QTL |
| 7 | QTL | 106146 | 8% | 4% | 105 | Search QTL | Search QTL |
| 8 | ENDOSPERM TRAIT | 98813 | 0% | 100% | 4 | Search ENDOSPERM+TRAIT | Search ENDOSPERM+TRAIT |
| 9 | OUTBRED POPULATION | 98809 | 0% | 67% | 6 | Search OUTBRED+POPULATION | Search OUTBRED+POPULATION |
| 10 | FUNCTIONAL MAPPING | 89737 | 2% | 14% | 26 | Search FUNCTIONAL+MAPPING | Search FUNCTIONAL+MAPPING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ZHANG, YM , (2006) ADVANCES ON METHODS FOR MAPPING QTL IN PLANT.CHINESE SCIENCE BULLETIN. VOL. 51. ISSUE 23. P. 2809-2818 | 59 | 60% | 7 |
| 2 | SILVA, LDE , ZENG, ZB , (2010) CURRENT PROGRESS ON STATISTICAL METHODS FOR MAPPING QUANTITATIVE TRAIT LOCI FROM INBRED LINE CROSSES.JOURNAL OF BIOPHARMACEUTICAL STATISTICS. VOL. 20. ISSUE 2. P. 454 -481 | 41 | 67% | 1 |
| 3 | SILLANPAA, MJ , HOTI, F , (2007) MAPPING QUANTITATIVE TRAIT LOCI FROM A SINGLE-TAIL SAMPLE OF THE PHENOTYPE DISTRIBUTION INCLUDING SURVIVAL DATA.GENETICS. VOL. 177. ISSUE 4. P. 2361-2377 | 43 | 62% | 6 |
| 4 | COFFMAN, CJ , DOERGE, RW , SIMONSEN, KL , NICHOLS, KM , DUARTE, C , WOLFINGER, RD , MCINTYRE, LM , (2005) MODEL SELECTION IN BINARY TRAIT LOCUS MAPPING.GENETICS. VOL. 170. ISSUE 3. P. 1281-1297 | 30 | 91% | 14 |
| 5 | HACKETT, CA , (2002) STATISTICAL METHODS FOR QTL MAPPING IN CEREALS.PLANT MOLECULAR BIOLOGY. VOL. 48. ISSUE 5. P. 585 -599 | 40 | 73% | 30 |
| 6 | HE, XH , QIN, HD , ZHANG, YM , ZHANG, TZ , HU, ZL , (2011) MAPPING OF EPISTATIC QUANTITATIVE TRAIT LOCI IN FOUR-WAY CROSSES.THEORETICAL AND APPLIED GENETICS. VOL. 122. ISSUE 1. P. 33-48 | 32 | 68% | 9 |
| 7 | LONDONO, D , CHEN, KM , MUSOLF, A , WANG, RX , SHEN, T , BRANDON, J , HERRING, JA , WISE, CA , ZOU, H , JIN, ML , ET AL (2013) A NOVEL METHOD FOR ANALYZING GENETIC ASSOCIATION WITH LONGITUDINAL PHENOTYPES.STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY. VOL. 12. ISSUE 2. P. 241-261 | 40 | 52% | 2 |
| 8 | LIU, YX , YANG, TF , LI, HW , YANG, RQ , (2014) ITERATIVELY REWEIGHTED LASSO FOR MAPPING MULTIPLE QUANTITATIVE TRAIT LOCI.BRIEFINGS IN BIOINFORMATICS. VOL. 15. ISSUE 1. P. 20-29 | 26 | 79% | 0 |
| 9 | YI, N , SHRINER, D , (2008) ADVANCES IN BAYESIAN MULTIPLE QUANTITATIVE TRAIT LOCI MAPPING IN EXPERIMENTAL CROSSES.HEREDITY. VOL. 100. ISSUE 3. P. 240-252 | 34 | 65% | 23 |
| 10 | GAZAFFI, R , MARGARIDO, GRA , PASTINA, MM , MOLLINARI, M , GARCIA, AAF , (2014) A MODEL FOR QUANTITATIVE TRAIT LOCI MAPPING, LINKAGE PHASE, AND SEGREGATION PATTERN ESTIMATION FOR A FULL-SIB PROGENY.TREE GENETICS & GENOMES. VOL. 10. ISSUE 4. P. 791 -801 | 31 | 63% | 3 |
Classes with closest relation at Level 1 |