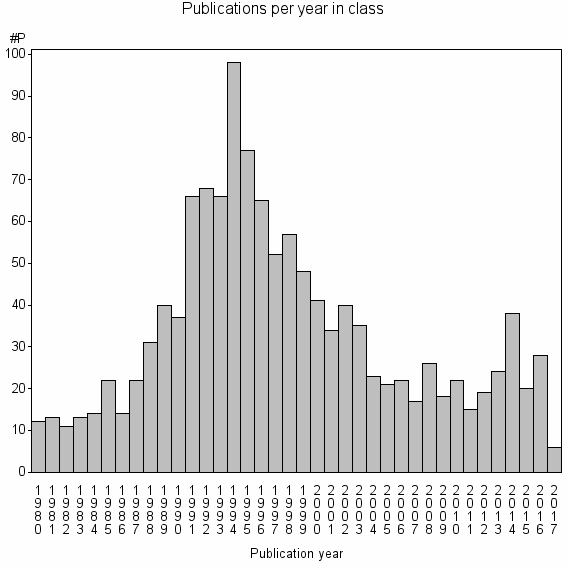
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 8251 | 1275 | 40.9 | 73% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 478 | 3 | PHAGE DISPLAY//CELL FREE PROTEIN SYNTHESIS//ANTIBODY ENGINEERING | 23687 |
| 1834 | 2 | PHAGE DISPLAY//ANTIBODY ENGINEERING//BISPECIFIC ANTIBODY | 6173 |
| 8251 | 1 | CANONICAL STRUCTURES//ANTIBODY MODELING//HUMANIZATION | 1275 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CANONICAL STRUCTURES | authKW | 367902 | 2% | 59% | 26 |
| 2 | ANTIBODY MODELING | authKW | 266176 | 1% | 65% | 17 |
| 3 | HUMANIZATION | authKW | 249425 | 3% | 27% | 39 |
| 4 | COMPLEMENTARITY DETERMINING REGION | authKW | 192393 | 2% | 27% | 30 |
| 5 | ANTIBODY STRUCTURE | authKW | 166656 | 2% | 30% | 23 |
| 6 | ANTIBODY ANTIGEN COMPLEX | authKW | 149881 | 1% | 48% | 13 |
| 7 | CDR CONFORMATION | authKW | 119738 | 0% | 100% | 5 |
| 8 | ANTIBODY HUMANIZATION | authKW | 107308 | 1% | 41% | 11 |
| 9 | HYPERVARIABLE LOOPS | authKW | 106671 | 1% | 64% | 7 |
| 10 | CDR H3 | authKW | 102169 | 1% | 53% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 10125 | 72% | 0% | 914 |
| 2 | Immunology | 2616 | 23% | 0% | 291 |
| 3 | Biophysics | 2216 | 16% | 0% | 202 |
| 4 | Biochemical Research Methods | 1292 | 11% | 0% | 145 |
| 5 | Biotechnology & Applied Microbiology | 148 | 6% | 0% | 77 |
| 6 | Crystallography | 130 | 3% | 0% | 43 |
| 7 | Mathematical & Computational Biology | 103 | 2% | 0% | 25 |
| 8 | Biology | 51 | 3% | 0% | 32 |
| 9 | Cell Biology | 39 | 5% | 0% | 64 |
| 10 | Computer Science, Interdisciplinary Applications | 6 | 1% | 0% | 16 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CRYSTALLOG PROGRAM | 49244 | 1% | 17% | 12 |
| 2 | HGEBIET PROT MODELLING | 47895 | 0% | 100% | 2 |
| 3 | 148 SIDNEY ST | 23948 | 0% | 100% | 1 |
| 4 | 1500 WALLACE BLVD | 23948 | 0% | 100% | 1 |
| 5 | 2375 GARCIA AVE | 23948 | 0% | 100% | 1 |
| 6 | ABT PROTE URFOR | 23948 | 0% | 100% | 1 |
| 7 | ANIM PHYSIOL GENET IMMUNOL | 23948 | 0% | 100% | 1 |
| 8 | ANTIBODY PREPARAT | 23948 | 0% | 100% | 1 |
| 9 | BASIC SCISTRUCT BIOPHYS | 23948 | 0% | 100% | 1 |
| 10 | BECKMAN INT PHYS BIOCHEM SECT | 23948 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR IMMUNOLOGY | 41167 | 9% | 2% | 110 |
| 2 | PROTEIN ENGINEERING | 24352 | 4% | 2% | 48 |
| 3 | JOURNAL OF MOLECULAR RECOGNITION | 24017 | 3% | 3% | 35 |
| 4 | JOURNAL OF MOLECULAR BIOLOGY | 15225 | 10% | 1% | 122 |
| 5 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 12558 | 5% | 1% | 58 |
| 6 | BIOTECHNOLOGY SOFTWARE & INTERNET JOURNAL | 11973 | 0% | 50% | 1 |
| 7 | BIOTECHNOLOGY SOFTWARE JOURNAL | 11973 | 0% | 50% | 1 |
| 8 | PROTEIN ENGINEERING DESIGN & SELECTION | 6634 | 1% | 2% | 17 |
| 9 | MABS | 5473 | 1% | 2% | 13 |
| 10 | PORTLAND PRESS PROCEEDINGS | 3984 | 0% | 4% | 4 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CANONICAL STRUCTURES | 367902 | 2% | 59% | 26 | Search CANONICAL+STRUCTURES | Search CANONICAL+STRUCTURES |
| 2 | ANTIBODY MODELING | 266176 | 1% | 65% | 17 | Search ANTIBODY+MODELING | Search ANTIBODY+MODELING |
| 3 | HUMANIZATION | 249425 | 3% | 27% | 39 | Search HUMANIZATION | Search HUMANIZATION |
| 4 | COMPLEMENTARITY DETERMINING REGION | 192393 | 2% | 27% | 30 | Search COMPLEMENTARITY+DETERMINING+REGION | Search COMPLEMENTARITY+DETERMINING+REGION |
| 5 | ANTIBODY STRUCTURE | 166656 | 2% | 30% | 23 | Search ANTIBODY+STRUCTURE | Search ANTIBODY+STRUCTURE |
| 6 | ANTIBODY ANTIGEN COMPLEX | 149881 | 1% | 48% | 13 | Search ANTIBODY+ANTIGEN+COMPLEX | Search ANTIBODY+ANTIGEN+COMPLEX |
| 7 | CDR CONFORMATION | 119738 | 0% | 100% | 5 | Search CDR+CONFORMATION | Search CDR+CONFORMATION |
| 8 | ANTIBODY HUMANIZATION | 107308 | 1% | 41% | 11 | Search ANTIBODY+HUMANIZATION | Search ANTIBODY+HUMANIZATION |
| 9 | HYPERVARIABLE LOOPS | 106671 | 1% | 64% | 7 | Search HYPERVARIABLE+LOOPS | Search HYPERVARIABLE+LOOPS |
| 10 | CDR H3 | 102169 | 1% | 53% | 8 | Search CDR+H3 | Search CDR+H3 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | PADLAN, EA , (1994) ANATOMY OF THE ANTIBODY MOLECULE.MOLECULAR IMMUNOLOGY. VOL. 31. ISSUE 3. P. 169-217 | 77 | 62% | 610 |
| 2 | PADLAN, EA , (1996) X-RAY CRYSTALLOGRAPHY OF ANTIBODIES.ADVANCES IN PROTEIN CHEMISTRY, VOL 49. VOL. 49. ISSUE . P. 57-133 | 72 | 65% | 119 |
| 3 | ALLAZIKANI, B , LESK, AM , CHOTHIA, C , (1997) STANDARD CONFORMATIONS FOR THE CANONICAL STRUCTURES OF IMMUNOGLOBULINS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 273. ISSUE 4. P. 927-948 | 40 | 82% | 399 |
| 4 | AKIBA, H , TSUMOTO, K , (2015) THERMODYNAMICS OF ANTIBODY-ANTIGEN INTERACTION REVEALED BY MUTATION ANALYSIS OF ANTIBODY VARIABLE REGIONS.JOURNAL OF BIOCHEMISTRY. VOL. 158. ISSUE 1. P. 1 -13 | 44 | 59% | 4 |
| 5 | KURODA, D , SHIRAI, H , JACOBSON, MP , NAKAMURA, H , (2012) COMPUTER-AIDED ANTIBODY DESIGN.PROTEIN ENGINEERING DESIGN & SELECTION. VOL. 25. ISSUE 10. P. 507-521 | 68 | 35% | 57 |
| 6 | MARCATILI, P , OLIMPIERI, PP , CHAILYAN, A , TRAMONTANO, A , (2014) ANTIBODY STRUCTURAL MODELING WITH PREDICTION OF IMMUNOGLOBULIN STRUCTURE (PIGS).NATURE PROTOCOLS. VOL. 9. ISSUE 12. P. 2771 -2783 | 43 | 60% | 7 |
| 7 | MOREA, V , TRAMONTANO, A , RUSTICI, M , CHOTHIA, C , LESK, AM , (1998) CONFORMATIONS OF THE THIRD HYPERVARIABLE REGION IN THE VH DOMAIN OF IMMUNOGLOBULINS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 275. ISSUE 2. P. 269 -294 | 47 | 65% | 227 |
| 8 | SWINDELLS, MB , PORTER, CT , COUCH, M , HURST, J , ABHINANDAN, KR , NIELSEN, JH , MACINDOE, G , HETHERINGTON, J , MARTIN, ACR , (2017) ABYSIS: INTEGRATED ANTIBODY SEQUENCE AND STRUCTURE-MANAGEMENT, ANALYSIS, AND PREDICTION.JOURNAL OF MOLECULAR BIOLOGY. VOL. 429. ISSUE 3. P. 356 -364 | 28 | 53% | 1 |
| 9 | SELA-CULANG, I , KUNIK, V , OFRAN, Y , (2013) THE STRUCTURAL BASIS OF ANTIBODY-ANTIGEN RECOGNITION.FRONTIERS IN IMMUNOLOGY. VOL. 4. ISSUE . P. - | 53 | 37% | 40 |
| 10 | MOHAN, S , SINHA, N , SMITH-GILL, J , (2003) MODELING THE BINDING SITES OF ANTI-HEN EGG WHITE LYSOZYME ANTIBODIES HYHEL-8 AND HYHEL-26: AN INSIGHT INTO THE MOLECULAR BASIS OF ANTIBODY CROSS-REACTIVITY AND SPECIFICITY.BIOPHYSICAL JOURNAL. VOL. 85. ISSUE 5. P. 3221 -3236 | 53 | 58% | 22 |
Classes with closest relation at Level 1 |