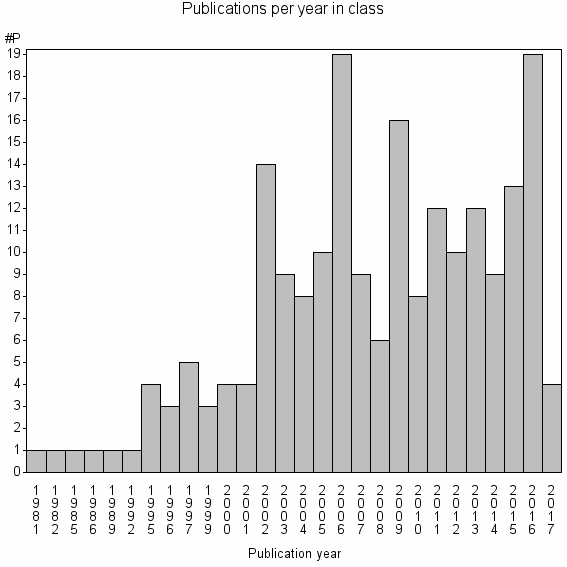
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 28425 | 207 | 42.5 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 28425 | 1 | PROCESSED PSEUDOGENES//CYP4Z2P//DUPLICATED PSEUDOGENES | 207 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROCESSED PSEUDOGENES | authKW | 451754 | 3% | 44% | 7 |
| 2 | CYP4Z2P | authKW | 295028 | 1% | 100% | 2 |
| 3 | DUPLICATED PSEUDOGENES | authKW | 295028 | 1% | 100% | 2 |
| 4 | PSEUDOGENES | authKW | 247886 | 10% | 8% | 20 |
| 5 | PSEUDOGENE | authKW | 230968 | 14% | 5% | 29 |
| 6 | RETROPSEUDOGENES | authKW | 196684 | 1% | 67% | 2 |
| 7 | CERNA | authKW | 157334 | 4% | 13% | 8 |
| 8 | ANTISENSERNA | authKW | 147514 | 0% | 100% | 1 |
| 9 | ARPAT | authKW | 147514 | 0% | 100% | 1 |
| 10 | BASS 432A | address | 147514 | 0% | 100% | 1 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 2998 | 50% | 0% | 103 |
| 2 | Biotechnology & Applied Microbiology | 687 | 25% | 0% | 52 |
| 3 | Evolutionary Biology | 506 | 11% | 0% | 22 |
| 4 | Biochemistry & Molecular Biology | 462 | 41% | 0% | 84 |
| 5 | Mathematical & Computational Biology | 222 | 6% | 0% | 13 |
| 6 | Cell Biology | 130 | 14% | 0% | 30 |
| 7 | Biochemical Research Methods | 54 | 6% | 0% | 13 |
| 8 | Oncology | 20 | 7% | 0% | 15 |
| 9 | Biology | 12 | 3% | 0% | 6 |
| 10 | Biophysics | 8 | 3% | 0% | 7 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BASS 432A | 147514 | 0% | 100% | 1 |
| 2 | COORDINAT COMPUTAT GENOM | 147514 | 0% | 100% | 1 |
| 3 | COUNCIL COMMUNIST YOUTH LEAGUE | 147514 | 0% | 100% | 1 |
| 4 | FRE 2326 DYNAM EVOLUT EXP S GENOMES MICRO | 147514 | 0% | 100% | 1 |
| 5 | JIANGSU CARCINOGENESIS INTRVENT | 147514 | 0% | 100% | 1 |
| 6 | LISHUI HOSPLISHUI MUNICIPAL CENT HOSP | 147514 | 0% | 100% | 1 |
| 7 | MOL BIOPHYS BICHEM | 147514 | 0% | 100% | 1 |
| 8 | RECH GENOM ENVIRONM | 147514 | 0% | 100% | 1 |
| 9 | ZHEJIANG UNIV THYROID BREAST SURG | 147514 | 0% | 100% | 1 |
| 10 | AFFILIATED NINGDE MUNICIPAL HOSP | 73756 | 0% | 50% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR BIOLOGY AND EVOLUTION | 4462 | 6% | 0% | 13 |
| 2 | BIOTECHFORUM ( BTF ) | 3640 | 4% | 0% | 8 |
| 3 | GENOME BIOLOGY | 3155 | 4% | 0% | 8 |
| 4 | BMC GENOMICS | 1530 | 5% | 0% | 10 |
| 5 | GENE | 909 | 5% | 0% | 11 |
| 6 | TRENDS IN GENETICS | 862 | 2% | 0% | 4 |
| 7 | ANNUAL REVIEW OF GENETICS | 711 | 1% | 0% | 2 |
| 8 | MAMMALIAN GENOME | 666 | 2% | 0% | 4 |
| 9 | NUCLEIC ACIDS RESEARCH | 615 | 6% | 0% | 13 |
| 10 | PLOS COMPUTATIONAL BIOLOGY | 504 | 2% | 0% | 4 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROCESSED PSEUDOGENES | 451754 | 3% | 44% | 7 | Search PROCESSED+PSEUDOGENES | Search PROCESSED+PSEUDOGENES |
| 2 | CYP4Z2P | 295028 | 1% | 100% | 2 | Search CYP4Z2P | Search CYP4Z2P |
| 3 | DUPLICATED PSEUDOGENES | 295028 | 1% | 100% | 2 | Search DUPLICATED+PSEUDOGENES | Search DUPLICATED+PSEUDOGENES |
| 4 | PSEUDOGENES | 247886 | 10% | 8% | 20 | Search PSEUDOGENES | Search PSEUDOGENES |
| 5 | PSEUDOGENE | 230968 | 14% | 5% | 29 | Search PSEUDOGENE | Search PSEUDOGENE |
| 6 | RETROPSEUDOGENES | 196684 | 1% | 67% | 2 | Search RETROPSEUDOGENES | Search RETROPSEUDOGENES |
| 7 | CERNA | 157334 | 4% | 13% | 8 | Search CERNA | Search CERNA |
| 8 | ANTISENSERNA | 147514 | 0% | 100% | 1 | Search ANTISENSERNA | Search ANTISENSERNA |
| 9 | ARPAT | 147514 | 0% | 100% | 1 | Search ARPAT | Search ARPAT |
| 10 | COMPETITIVE ENDOGENOUS RNAS | 147514 | 0% | 100% | 1 | Search COMPETITIVE+ENDOGENOUS+RNAS | Search COMPETITIVE+ENDOGENOUS+RNAS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LI, W , YANG, W , WANG, XJ , (2013) PSEUDOGENES: PSEUDO OR REAL FUNCTIONAL ELEMENTS?.JOURNAL OF GENETICS AND GENOMICS. VOL. 40. ISSUE 4. P. 171-177 | 40 | 60% | 21 |
| 2 | LIU, GQ , BAIYINBAOLIGAO , XING, YQ , (2010) ADVANCES IN RESEARCH ON PSEUDOGENES.PROGRESS IN BIOCHEMISTRY AND BIOPHYSICS. VOL. 37. ISSUE 11. P. 1165-1174 | 34 | 59% | 1 |
| 3 | CALEY, DP , PINK, RC , WICKS, K , PUNCH, EK , JACOBS, L , CARTER, DRF , (2011) PSEUDOGENES: PSEUDO-FUNCTIONAL OR KEY REGULATORS IN HEALTH AND DISEASE?.RNA. VOL. 17. ISSUE 5. P. 792 -798 | 32 | 43% | 96 |
| 4 | ZHENG, DY , GERSTEIN, MB , (2007) THE AMBIGUOUS BOUNDARY BETWEEN GENES AND PSEUDOGENES: THE DEAD RISE UP, OR DO THEY?.TRENDS IN GENETICS. VOL. 23. ISSUE 5. P. 219 -224 | 27 | 60% | 60 |
| 5 | SEN, K , GHOSH, TC , (2013) PSEUDOGENES AND THEIR COMPOSERS: DELVING IN THE 'DEBRIS' OF HUMAN GENOME.BRIEFINGS IN FUNCTIONAL GENOMICS. VOL. 12. ISSUE 6. P. 536-547 | 36 | 41% | 4 |
| 6 | PEI, BK , SISU, C , FRANKISH, A , HOWALD, C , HABEGGER, L , MU, XJ , HARTE, R , BALASUBRAMANIAN, S , TANZER, A , DIEKHANS, M , ET AL (2012) THE GENCODE PSEUDOGENE RESOURCE.GENOME BIOLOGY. VOL. 13. ISSUE 9. P. - | 24 | 47% | 90 |
| 7 | PINK, RC , CARTER, DRF , (2013) PSEUDOGENES AS REGULATORS OF BIOLOGICAL FUNCTION.ROLE OF NON-CODING RNAS IN BIOLOGY. VOL. 54. ISSUE . P. 103 -112 | 24 | 60% | 7 |
| 8 | XIAO, J , SEKHWAL, MK , LI, PC , RAGUPATHY, R , CLOUTIER, S , WANG, X , YOU, FM , (2016) PSEUDOGENES AND THEIR GENOME-WIDE PREDICTION IN PLANTS.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 17. ISSUE 12. P. - | 29 | 38% | 0 |
| 9 | GUO, XY , LIN, MY , ROCKOWITZ, S , LACHMAN, HM , ZHENG, DY , (2014) CHARACTERIZATION OF HUMAN PSEUDOGENE- DERIVED NON- CODING RNAS FOR FUNCTIONAL POTENTIAL.PLOS ONE. VOL. 9. ISSUE 4. P. - | 31 | 33% | 10 |
| 10 | ROUCHKA, EC , CHA, IE , (2009) CURRENT TRENDS IN PSEUDOGENE DETECTION AND CHARACTERIZATION.CURRENT BIOINFORMATICS. VOL. 4. ISSUE 2. P. 112-119 | 31 | 39% | 9 |
Classes with closest relation at Level 1 |