Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
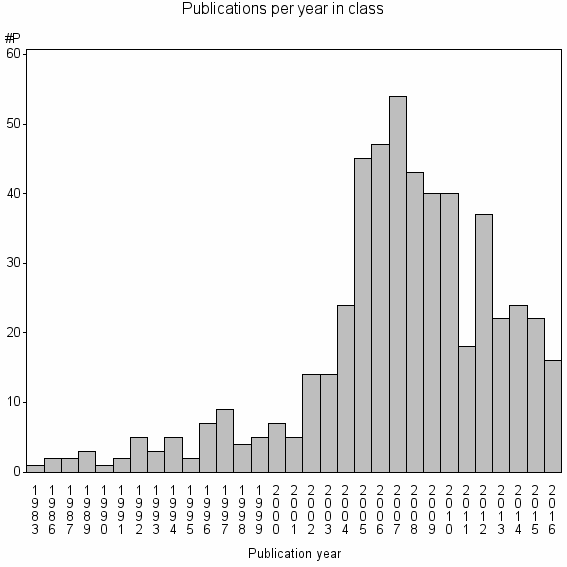
| 19074 | 523 | 41.9 | 88% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 126 | 2 | MOLECULAR BIOLOGY AND EVOLUTION//BIOINFORMATICS//MATHEMATICAL & COMPUTATIONAL BIOLOGY | 23963 |
| 19074 | 1 | NATURAL ANTISENSE TRANSCRIPT//WRAP53//OVERLAPPING GENES | 523 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | NATURAL ANTISENSE TRANSCRIPT | authKW | 451680 | 4% | 37% | 21 |
| 2 | WRAP53 | authKW | 449102 | 2% | 77% | 10 |
| 3 | OVERLAPPING GENES | authKW | 328895 | 4% | 28% | 20 |
| 4 | CUSTOM BIOTECHNOL SERV GRP | address | 233536 | 1% | 100% | 4 |
| 5 | ENDOGENOUS ANTISENSE RNA | authKW | 175152 | 1% | 100% | 3 |
| 6 | GENOME IL | address | 168275 | 1% | 41% | 7 |
| 7 | TILING ARRAY | authKW | 164428 | 2% | 22% | 13 |
| 8 | HYBRID RNA | authKW | 131362 | 1% | 75% | 3 |
| 9 | NUDT6 | authKW | 131362 | 1% | 75% | 3 |
| 10 | TECHNOL DEV TEAM MAMMALIAN CELLULAR DYNAM | address | 127800 | 2% | 24% | 9 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 4384 | 38% | 0% | 200 |
| 2 | Mathematical & Computational Biology | 4019 | 16% | 0% | 85 |
| 3 | Biotechnology & Applied Microbiology | 3191 | 33% | 0% | 175 |
| 4 | Biochemical Research Methods | 1093 | 16% | 0% | 83 |
| 5 | Biochemistry & Molecular Biology | 1076 | 39% | 0% | 205 |
| 6 | Cell Biology | 310 | 14% | 0% | 74 |
| 7 | Evolutionary Biology | 213 | 5% | 0% | 24 |
| 8 | Statistics & Probability | 85 | 4% | 0% | 19 |
| 9 | Biology | 72 | 4% | 0% | 21 |
| 10 | Virology | 35 | 2% | 0% | 13 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CUSTOM BIOTECHNOL SERV GRP | 233536 | 1% | 100% | 4 |
| 2 | GENOME IL | 168275 | 1% | 41% | 7 |
| 3 | TECHNOL DEV TEAM MAMMALIAN CELLULAR DYNAM | 127800 | 2% | 24% | 9 |
| 4 | LEHRSTUHL DATENANAL VISUALISIERUNG | 116768 | 0% | 100% | 2 |
| 5 | ARABIDOPSIS THALIANA GENOME | 58384 | 0% | 100% | 1 |
| 6 | AUSTRALIA AUSTRALIAN CANC STUDV | 58384 | 0% | 100% | 1 |
| 7 | BIOINFORMAT PEKING YALE JOINT PLANT | 58384 | 0% | 100% | 1 |
| 8 | COMPART GENOM BIOINFORMAT | 58384 | 0% | 100% | 1 |
| 9 | CURIEUMR3244 | 58384 | 0% | 100% | 1 |
| 10 | DEV BIOLDEV VASC BIOL PROGRAM | 58384 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOTECHFORUM ( BTF ) | 8125 | 4% | 1% | 19 |
| 2 | BIOINFORMATICS | 7070 | 7% | 0% | 36 |
| 3 | GENOME BIOLOGY | 4989 | 3% | 1% | 16 |
| 4 | BMC BIOINFORMATICS | 4086 | 4% | 0% | 23 |
| 5 | BMC GENOMICS | 3784 | 5% | 0% | 25 |
| 6 | TRENDS IN GENETICS | 2133 | 2% | 0% | 10 |
| 7 | STATISTICAL APPLICATIONS IN GENETICS AND MOLECULAR BIOLOGY | 1842 | 1% | 1% | 4 |
| 8 | BIOLOGY DIRECT | 1678 | 1% | 1% | 4 |
| 9 | BMC MOLECULAR BIOLOGY | 1226 | 1% | 1% | 4 |
| 10 | NUCLEIC ACIDS RESEARCH | 1121 | 5% | 0% | 28 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NATURAL ANTISENSE TRANSCRIPT | 451680 | 4% | 37% | 21 | Search NATURAL+ANTISENSE+TRANSCRIPT | Search NATURAL+ANTISENSE+TRANSCRIPT |
| 2 | WRAP53 | 449102 | 2% | 77% | 10 | Search WRAP53 | Search WRAP53 |
| 3 | OVERLAPPING GENES | 328895 | 4% | 28% | 20 | Search OVERLAPPING+GENES | Search OVERLAPPING+GENES |
| 4 | ENDOGENOUS ANTISENSE RNA | 175152 | 1% | 100% | 3 | Search ENDOGENOUS+ANTISENSE+RNA | Search ENDOGENOUS+ANTISENSE+RNA |
| 5 | TILING ARRAY | 164428 | 2% | 22% | 13 | Search TILING+ARRAY | Search TILING+ARRAY |
| 6 | HYBRID RNA | 131362 | 1% | 75% | 3 | Search HYBRID+RNA | Search HYBRID+RNA |
| 7 | NUDT6 | 131362 | 1% | 75% | 3 | Search NUDT6 | Search NUDT6 |
| 8 | AFFYMETRIX TILING ARRAYS | 116768 | 0% | 100% | 2 | Search AFFYMETRIX+TILING+ARRAYS | Search AFFYMETRIX+TILING+ARRAYS |
| 9 | ANTISENSE TRANSCRIPTOME | 116768 | 0% | 100% | 2 | Search ANTISENSE+TRANSCRIPTOME | Search ANTISENSE+TRANSCRIPTOME |
| 10 | LONG NATURALLY OCCURRING ANTISENSE TRANSCRIPTS NATS | 116768 | 0% | 100% | 2 | Search LONG+NATURALLY+OCCURRING+ANTISENSE+TRANSCRIPTS+NATS | Search LONG+NATURALLY+OCCURRING+ANTISENSE+TRANSCRIPTS+NATS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ROSIKIEWICZ, W , MAKALOWSKA, I , (2016) BIOLOGICAL FUNCTIONS OF NATURAL ANTISENSE TRANSCRIPTS.ACTA BIOCHIMICA POLONICA. VOL. 63. ISSUE 4. P. 665 -673 | 45 | 39% | 0 |
| 2 | CHEREZOV, RO , SIMONOVA, OB , (2014) OVERLAPPING GENES AND ANTISENSE TRANSCRIPTION IN EUKARYOTES.RUSSIAN JOURNAL OF GENETICS. VOL. 50. ISSUE 7. P. 653 -666 | 46 | 39% | 1 |
| 3 | HO, MR , TSAI, KW , LIN, WC , (2012) A UNIFIED FRAMEWORK OF OVERLAPPING GENES: TOWARDS THE ORIGINATION AND ENDOGENIC REGULATION.GENOMICS. VOL. 100. ISSUE 4. P. 231 -239 | 28 | 54% | 10 |
| 4 | FONSECA, MM , HARRIS, DJ , POSADA, D , (2014) ORIGIN AND LENGTH DISTRIBUTION OF UNIDIRECTIONAL PROKARYOTIC OVERLAPPING GENES.G3-GENES GENOMES GENETICS. VOL. 4. ISSUE 1. P. 19-27 | 18 | 72% | 5 |
| 5 | BEITER, T , REICH, E , WILLIAMS, RW , SIMON, P , (2009) ANTISENSE TRANSCRIPTION: A CRITICAL LOOK IN BOTH DIRECTIONS.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 66. ISSUE 1. P. 94 -112 | 53 | 25% | 61 |
| 6 | BRITTO-KIDO, SD , NETO, JRCF , PANDOLFI, V , MARCELINO-GUIMARAES, FC , NEPOMUCENO, AL , ABDELNOOR, RV , BENKO-ISEPPON, A , KIDO, EA , (2013) NATURAL ANTISENSE TRANSCRIPTS IN PLANTS: A REVIEW AND IDENTIFICATION IN SOYBEAN INFECTED WITH PHAKOPSORA PACHYRHIZI SUPERSAGE LIBRARY.SCIENTIFIC WORLD JOURNAL. VOL. . ISSUE . P. - | 27 | 47% | 1 |
| 7 | ZHANG, YC , LIN, K , (2015) PHYLOGENY INFERENCE OF CLOSELY RELATED BACTERIAL GENOMES: COMBINING THE FEATURES OF BOTH OVERLAPPING GENES AND COLLINEAR GENOMIC REGIONS.EVOLUTIONARY BIOINFORMATICS. VOL. 11. ISSUE . P. 1 -9 | 22 | 48% | 0 |
| 8 | NUMATA, K , KIYOSAWA, H , (2012) GENOME-WIDE IMPACT OF ENDOGENOUS ANTISENSE TRANSCRIPTS IN EUKARYOTES.FRONTIERS IN BIOSCIENCE-LANDMARK. VOL. 17. ISSUE . P. 300-315 | 37 | 29% | 4 |
| 9 | SOLDA, G , SUYAMA, M , PELUCCHI, P , BOI, S , GUFFANTI, A , RIZZI, E , BORK, P , TENCHINI, ML , CICCARELLI, FD , (2008) NON-RANDOM RETENTION OF PROTEIN-CODING OVERLAPPING GENES IN METAZOA.BMC GENOMICS. VOL. 9. ISSUE . P. - | 24 | 49% | 13 |
| 10 | MAKALOWSKA, I , LIN, CF , MAKALOWSKI, W , (2005) OVERLAPPING GENES IN VERTEBRATE GENOMES.COMPUTATIONAL BIOLOGY AND CHEMISTRY. VOL. 29. ISSUE 1. P. 1-12 | 32 | 36% | 54 |
Classes with closest relation at Level 1 |