Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
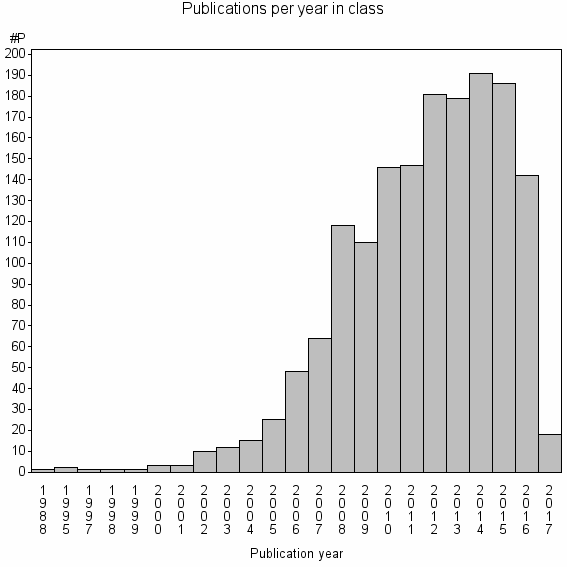
| 5569 | 1604 | 44.9 | 91% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 1101 | 2 | BIOINFORMATICS//COPY NUMBER VARIATION//NEXT GENERATION SEQUENCING | 9808 |
| 5569 | 1 | COPY NUMBER VARIATION//COPY NUMBER VARIANTS//STRUCTURAL VARIATION | 1604 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | COPY NUMBER VARIATION | authKW | 1044027 | 15% | 23% | 234 |
| 2 | COPY NUMBER VARIANTS | authKW | 244623 | 4% | 21% | 62 |
| 3 | STRUCTURAL VARIATION | authKW | 225282 | 3% | 29% | 41 |
| 4 | CNV | authKW | 214723 | 5% | 14% | 80 |
| 5 | PENNCNV | authKW | 191937 | 1% | 92% | 11 |
| 6 | COPY NUMBER VARIATION CNV | authKW | 141795 | 1% | 32% | 23 |
| 7 | CNVS | authKW | 116444 | 1% | 32% | 19 |
| 8 | CCL3L1 | authKW | 96358 | 1% | 56% | 9 |
| 9 | NBPF | authKW | 79312 | 0% | 83% | 5 |
| 10 | PARALOGUE RATIO TEST | authKW | 76141 | 0% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Genetics & Heredity | 36727 | 62% | 0% | 997 |
| 2 | Mathematical & Computational Biology | 7090 | 12% | 0% | 199 |
| 3 | Biotechnology & Applied Microbiology | 6966 | 28% | 0% | 457 |
| 4 | Biochemical Research Methods | 2352 | 13% | 0% | 216 |
| 5 | Biochemistry & Molecular Biology | 676 | 21% | 0% | 333 |
| 6 | Psychiatry | 238 | 5% | 0% | 88 |
| 7 | Agriculture, Dairy & Animal Science | 61 | 2% | 0% | 31 |
| 8 | Evolutionary Biology | 48 | 2% | 0% | 25 |
| 9 | Cell Biology | 24 | 4% | 0% | 68 |
| 10 | Statistics & Probability | 24 | 1% | 0% | 24 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CELLULAR MOL MOL MED | 57106 | 0% | 100% | 3 |
| 2 | GRP MOL GENET SYST BIOL | 57106 | 0% | 100% | 3 |
| 3 | QUANTITAT GENOM MED | 50758 | 0% | 67% | 4 |
| 4 | PL GENOM | 50477 | 3% | 5% | 50 |
| 5 | GENOME SCI | 44570 | 7% | 2% | 110 |
| 6 | SHANDONG PROV ANIM DIS CONTROL BREEDING | 40302 | 0% | 35% | 6 |
| 7 | BIOINFORMAT BIOFOTON | 38071 | 0% | 100% | 2 |
| 8 | EPIDEMIOL PUBL HLTH MD3 | 38071 | 0% | 100% | 2 |
| 9 | GEL BOVINE FUNCT GENOM | 38071 | 0% | 100% | 2 |
| 10 | SECCIO GENET CLIN | 38071 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC GENOMICS | 22686 | 7% | 1% | 107 |
| 2 | GENOME RESEARCH | 18894 | 2% | 3% | 37 |
| 3 | BIOINFORMATICS | 14698 | 6% | 1% | 91 |
| 4 | NATURE GENETICS | 11998 | 3% | 1% | 56 |
| 5 | BIOTECHFORUM ( BTF ) | 11137 | 2% | 2% | 39 |
| 6 | BMC BIOINFORMATICS | 6522 | 3% | 1% | 51 |
| 7 | CYTOGENETIC AND GENOME RESEARCH | 6353 | 2% | 1% | 27 |
| 8 | HUMAN MUTATION | 5494 | 2% | 1% | 34 |
| 9 | AMERICAN JOURNAL OF HUMAN GENETICS | 5169 | 3% | 1% | 47 |
| 10 | GENOME BIOLOGY | 4958 | 2% | 1% | 28 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | COPY NUMBER VARIATION | 1044027 | 15% | 23% | 234 | Search COPY+NUMBER+VARIATION | Search COPY+NUMBER+VARIATION |
| 2 | COPY NUMBER VARIANTS | 244623 | 4% | 21% | 62 | Search COPY+NUMBER+VARIANTS | Search COPY+NUMBER+VARIANTS |
| 3 | STRUCTURAL VARIATION | 225282 | 3% | 29% | 41 | Search STRUCTURAL+VARIATION | Search STRUCTURAL+VARIATION |
| 4 | CNV | 214723 | 5% | 14% | 80 | Search CNV | Search CNV |
| 5 | PENNCNV | 191937 | 1% | 92% | 11 | Search PENNCNV | Search PENNCNV |
| 6 | COPY NUMBER VARIATION CNV | 141795 | 1% | 32% | 23 | Search COPY+NUMBER+VARIATION+CNV | Search COPY+NUMBER+VARIATION+CNV |
| 7 | CNVS | 116444 | 1% | 32% | 19 | Search CNVS | Search CNVS |
| 8 | CCL3L1 | 96358 | 1% | 56% | 9 | Search CCL3L1 | Search CCL3L1 |
| 9 | NBPF | 79312 | 0% | 83% | 5 | Search NBPF | Search NBPF |
| 10 | PARALOGUE RATIO TEST | 76141 | 0% | 100% | 4 | Search PARALOGUE+RATIO+TEST | Search PARALOGUE+RATIO+TEST |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ZANDI, PP , PIROOZNIA, M , GOES, FS , (2015) WHOLE-GENOME CNV ANALYSIS: ADVANCES IN COMPUTATIONAL APPROACHES.FRONTIERS IN GENETICS. VOL. 6. ISSUE . P. - | 60 | 88% | 8 |
| 2 | ESCARAMIS, G , DOCAMPO, E , RABIONET, R , (2015) A DECADE OF STRUCTURAL VARIANTS: DESCRIPTION, HISTORY AND METHODS TO DETECT STRUCTURAL VARIATION.BRIEFINGS IN FUNCTIONAL GENOMICS. VOL. 14. ISSUE 5. P. 305 -314 | 81 | 68% | 4 |
| 3 | ALKAN, C , COE, BP , EICHLER, EE , (2011) APPLICATIONS OF NEXT-GENERATION SEQUENCING GENOME STRUCTURAL VARIATION DISCOVERY AND GENOTYPING.NATURE REVIEWS GENETICS. VOL. 12. ISSUE 5. P. 363 -375 | 57 | 54% | 371 |
| 4 | MILLS, RE , WALTER, K , STEWART, C , HANDSAKER, RE , CHEN, K , ALKAN, C , ABYZOV, A , YOON, SC , YE, K , CHEETHAM, RK , ET AL (2011) MAPPING COPY NUMBER VARIATION BY POPULATION-SCALE GENOME SEQUENCING.NATURE. VOL. 470. ISSUE 7332. P. 59-65 | 33 | 80% | 392 |
| 5 | LIU, GE , BICKHART, DM , (2012) COPY NUMBER VARIATION IN THE CATTLE GENOME.FUNCTIONAL & INTEGRATIVE GENOMICS. VOL. 12. ISSUE 4. P. 609 -624 | 84 | 57% | 17 |
| 6 | ZHAO, M , WANG, QG , WANG, Q , JIA, PL , ZHAO, ZM , (2013) COMPUTATIONAL TOOLS FOR COPY NUMBER VARIATION (CNV) DETECTION USING NEXT-GENERATION SEQUENCING DATA: FEATURES AND PERSPECTIVES.BMC BIOINFORMATICS. VOL. 14. ISSUE . P. - | 52 | 71% | 55 |
| 7 | ZARREI, M , MACDONALD, JR , MERICO, D , SCHERER, SW , (2015) A COPY NUMBER VARIATION MAP OF THE HUMAN GENOME.NATURE REVIEWS GENETICS. VOL. 16. ISSUE 3. P. 172 -183 | 47 | 52% | 64 |
| 8 | GIRIRAJAN, S , CAMPBELL, CD , EICHLER, EE , (2011) HUMAN COPY NUMBER VARIATION AND COMPLEX GENETIC DISEASE.ANNUAL REVIEW OF GENETICS, VOL 45. VOL. 45. ISSUE . P. 203 -226 | 66 | 51% | 110 |
| 9 | CARVALHO, CMB , LUPSKI, JR , (2016) MECHANISMS UNDERLYING STRUCTURAL VARIANT FORMATION IN GENOMIC DISORDERS.NATURE REVIEWS GENETICS. VOL. 17. ISSUE 4. P. 224 -238 | 59 | 38% | 12 |
| 10 | JENKINS, GM , GODDARD, ME , BLACK, MA , BRAUNING, R , AUVRAY, B , DODDS, KG , KIJAS, JW , COCKETT, N , MCEWAN, JC , (2016) COPY NUMBER VARIANTS IN THE SHEEP GENOME DETECTED USING MULTIPLE APPROACHES.BMC GENOMICS. VOL. 17. ISSUE . P. - | 50 | 76% | 0 |
Classes with closest relation at Level 1 |