Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
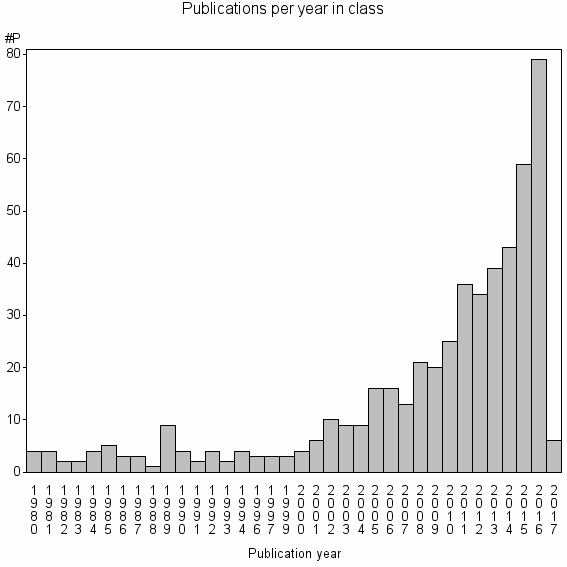
| 19394 | 507 | 48.9 | 86% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 262 | 3 | AUTOPHAGY//RESVERATROL//HISTONE DEACETYLASE INHIBITOR | 44558 |
| 2649 | 2 | SIRT1//SIRTUINS//SIRT3 | 3472 |
| 19394 | 1 | N TERMINAL ACETYLTRANSFERASE//ARD1//N TERMINAL ACETYLATION | 507 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | N TERMINAL ACETYLTRANSFERASE | authKW | 619469 | 2% | 86% | 12 |
| 2 | ARD1 | authKW | 615943 | 3% | 68% | 15 |
| 3 | N TERMINAL ACETYLATION | authKW | 510742 | 4% | 40% | 21 |
| 4 | LYSINE ACETYLATION | authKW | 452122 | 6% | 24% | 31 |
| 5 | NAA10 | authKW | 361359 | 1% | 100% | 6 |
| 6 | NAA10P | authKW | 301132 | 1% | 100% | 5 |
| 7 | PROTEIN N TERMINAL ACETYLATION | authKW | 301132 | 1% | 100% | 5 |
| 8 | ACETYLOME | authKW | 256745 | 2% | 47% | 9 |
| 9 | LYSINE SUCCINYLATION | authKW | 245919 | 1% | 58% | 7 |
| 10 | HARD1 | authKW | 240906 | 1% | 100% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 2400 | 57% | 0% | 287 |
| 2 | Biochemical Research Methods | 1940 | 21% | 0% | 107 |
| 3 | Cell Biology | 513 | 18% | 0% | 90 |
| 4 | Microbiology | 267 | 11% | 0% | 55 |
| 5 | Biophysics | 172 | 8% | 0% | 39 |
| 6 | Genetics & Heredity | 110 | 7% | 0% | 38 |
| 7 | Oncology | 42 | 7% | 0% | 35 |
| 8 | Biotechnology & Applied Microbiology | 33 | 5% | 0% | 25 |
| 9 | Mycology | 33 | 1% | 0% | 6 |
| 10 | Developmental Biology | 8 | 1% | 0% | 6 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CHEM DEF MICROSCALE ANAL | 120453 | 0% | 100% | 2 |
| 2 | PROGRAM GENE EXP S REGULAT | 95186 | 1% | 23% | 7 |
| 3 | PISSARO PROTE IL | 74119 | 1% | 31% | 4 |
| 4 | CAROLINA BIOCHEM BIOPHYS | 60226 | 0% | 100% | 1 |
| 5 | CEA CHIM PROT | 60226 | 0% | 100% | 1 |
| 6 | CHILDRENS HOSP FDN HEMATOL ONCOL | 60226 | 0% | 100% | 1 |
| 7 | ECOENVIRONM THREE GORGES ERVOIR REGTH | 60226 | 0% | 100% | 1 |
| 8 | EMMAS CHILDREN HOSP GENET METAB DIS | 60226 | 0% | 100% | 1 |
| 9 | ESTUDIOS INVEST TECN TECNUN | 60226 | 0% | 100% | 1 |
| 10 | HANGZHOU ECO TECH DEV AREA | 60226 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR & CELLULAR PROTEOMICS | 9436 | 4% | 1% | 22 |
| 2 | JOURNAL OF PROTEOME RESEARCH | 5663 | 5% | 0% | 24 |
| 3 | PROTEOMICS | 3777 | 4% | 0% | 19 |
| 4 | JOURNAL OF PROTEOMICS | 2749 | 2% | 0% | 11 |
| 5 | MOLECULAR SYSTEMS BIOLOGY | 1267 | 1% | 1% | 4 |
| 6 | JOURNAL OF BIOLOGICAL CHEMISTRY | 822 | 9% | 0% | 47 |
| 7 | MICROBIOLOGYOPEN | 680 | 0% | 1% | 2 |
| 8 | PLANT SIGNALING & BEHAVIOR | 558 | 0% | 0% | 2 |
| 9 | CURRENT OPINION IN MICROBIOLOGY | 520 | 1% | 0% | 4 |
| 10 | MOLECULAR MICROBIOLOGY | 491 | 2% | 0% | 10 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | N TERMINAL ACETYLTRANSFERASE | 619469 | 2% | 86% | 12 | Search N+TERMINAL+ACETYLTRANSFERASE | Search N+TERMINAL+ACETYLTRANSFERASE |
| 2 | ARD1 | 615943 | 3% | 68% | 15 | Search ARD1 | Search ARD1 |
| 3 | N TERMINAL ACETYLATION | 510742 | 4% | 40% | 21 | Search N+TERMINAL+ACETYLATION | Search N+TERMINAL+ACETYLATION |
| 4 | LYSINE ACETYLATION | 452122 | 6% | 24% | 31 | Search LYSINE+ACETYLATION | Search LYSINE+ACETYLATION |
| 5 | NAA10 | 361359 | 1% | 100% | 6 | Search NAA10 | Search NAA10 |
| 6 | NAA10P | 301132 | 1% | 100% | 5 | Search NAA10P | Search NAA10P |
| 7 | PROTEIN N TERMINAL ACETYLATION | 301132 | 1% | 100% | 5 | Search PROTEIN+N+TERMINAL+ACETYLATION | Search PROTEIN+N+TERMINAL+ACETYLATION |
| 8 | ACETYLOME | 256745 | 2% | 47% | 9 | Search ACETYLOME | Search ACETYLOME |
| 9 | LYSINE SUCCINYLATION | 245919 | 1% | 58% | 7 | Search LYSINE+SUCCINYLATION | Search LYSINE+SUCCINYLATION |
| 10 | HARD1 | 240906 | 1% | 100% | 4 | Search HARD1 | Search HARD1 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | DORFEL, MJ , LYON, GJ , (2015) THE BIOLOGICAL FUNCTIONS OF NAA10-FROM AMINO-TERMINAL ACETYLATION TO HUMAN DISEASE.GENE. VOL. 567. ISSUE 2. P. 103 -131 | 123 | 44% | 10 |
| 2 | STARHEIM, KK , GEVAERT, K , ARNESEN, T , (2012) PROTEIN N-TERMINAL ACETYLTRANSFERASES: WHEN THE START MATTERS.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 37. ISSUE 4. P. 152 -161 | 51 | 84% | 93 |
| 3 | AKSNES, H , DRAZIC, A , MARIE, M , ARNESEN, T , (2016) FIRST THINGS FIRST: VITAL PROTEIN VARKS BY N-TERMINAL ACETYLTRANSFERASES.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 41. ISSUE 9. P. 746 -760 | 64 | 73% | 1 |
| 4 | RATHORE, OS , FAUSTINO, A , PRUDENCIO, P , VAN DAMME, P , COX, CJ , MARTINHO, RG , (2016) ABSENCE OF N-TERMINAL ACETYLTRANSFERASE DIVERSIFICATION DURING EVOLUTION OF EUKARYOTIC ORGANISMS.SCIENTIFIC REPORTS. VOL. 6. ISSUE . P. - | 55 | 67% | 3 |
| 5 | KALVIK, TV , ARNESEN, T , (2013) PROTEIN N-TERMINAL ACETYLTRANSFERASES IN CANCER.ONCOGENE. VOL. 32. ISSUE 3. P. 269-276 | 54 | 68% | 37 |
| 6 | BERNAL, V , CASTANO-CEREZO, S , GALLEGO-JARA, J , ECIJA-CONESA, A , DE DIEGO, T , IBORRA, JL , CANOVAS, M , (2014) REGULATION OF BACTERIAL PHYSIOLOGY BY LYSINE ACETYLATION OF PROTEINS.NEW BIOTECHNOLOGY. VOL. 31. ISSUE 6. P. 586 -595 | 48 | 64% | 19 |
| 7 | STOVE, SI , MAGIN, RS , FOYN, H , HAUG, BE , MARMORSTEIN, R , ARNESEN, T , (2016) CRYSTAL STRUCTURE OF THE GOLGI-ASSOCIATED HUMAN N ALPHA-ACETYLTRANSFERASE 60 REVEALS THE MOLECULAR DETERMINANTS FOR SUBSTRATE-SPECIFIC ACETYLATION.STRUCTURE. VOL. 24. ISSUE 7. P. 1044 -1056 | 45 | 76% | 0 |
| 8 | DRAZIC, A , MYKLEBUST, LM , REE, R , ARNESEN, T , (2016) THE WORLD OF PROTEIN ACETYLATION.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1864. ISSUE 10. P. 1372 -1401 | 100 | 24% | 4 |
| 9 | OUIDIR, T , KENTACHE, T , HARDOUIN, J , (2016) PROTEIN LYSINE ACETYLATION IN BACTERIA: CURRENT STATE OF THE ART.PROTEOMICS. VOL. 16. ISSUE 2. P. 301 -309 | 42 | 59% | 3 |
| 10 | WOLFE, AJ , (2016) BACTERIAL PROTEIN ACETYLATION: NEW DISCOVERIES UNANSWERED QUESTIONS.CURRENT GENETICS. VOL. 62. ISSUE 2. P. 335 -341 | 42 | 56% | 4 |
Classes with closest relation at Level 1 |